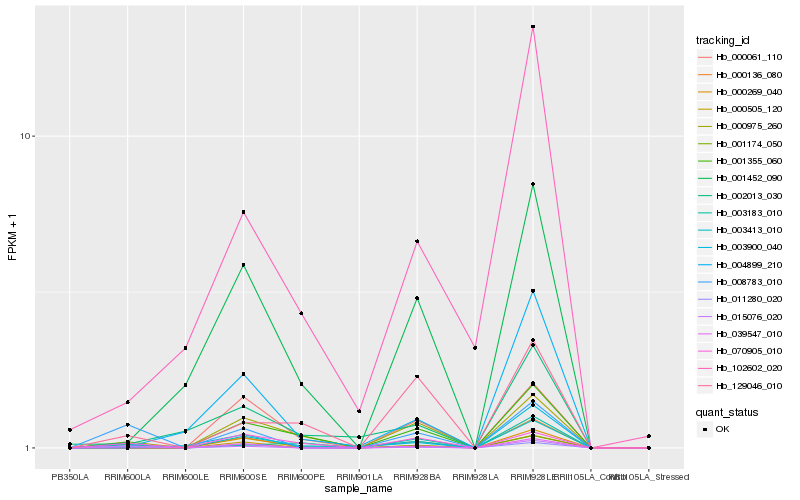
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003413_010 |
0.0 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 2 |
Hb_000136_080 |
0.1279740372 |
- |
- |
hypothetical protein AALP_AA8G499800 [Arabis alpina] |
| 3 |
Hb_000975_260 |
0.1726283438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000505_120 |
0.1957851104 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 5 |
Hb_011280_020 |
0.2097877233 |
- |
- |
polyprotein [Oryza australiensis] |
| 6 |
Hb_070905_010 |
0.2234175157 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 7 |
Hb_001174_050 |
0.226923835 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_001355_060 |
0.2366948557 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 9 |
Hb_015076_020 |
0.2426807121 |
- |
- |
IMP dehydrogenase/GMP reductase [Hevea brasiliensis] |
| 10 |
Hb_008783_010 |
0.242870036 |
- |
- |
- |
| 11 |
Hb_002013_030 |
0.2448418021 |
- |
- |
PREDICTED: uncharacterized protein LOC104799692 [Tarenaya hassleriana] |
| 12 |
Hb_000269_040 |
0.2500235786 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_004899_210 |
0.2502869953 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein E [Jatropha curcas] |
| 14 |
Hb_000061_110 |
0.2506528697 |
- |
- |
- |
| 15 |
Hb_102602_020 |
0.2513261153 |
- |
- |
hypothetical protein B456_001G166300 [Gossypium raimondii] |
| 16 |
Hb_003183_010 |
0.257154704 |
- |
- |
PREDICTED: uncharacterized protein LOC105785048, partial [Gossypium raimondii] |
| 17 |
Hb_003900_040 |
0.2580973781 |
- |
- |
hypothetical protein [Lotus japonicus] |
| 18 |
Hb_001452_090 |
0.2586952416 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Vitis vinifera] |
| 19 |
Hb_129046_010 |
0.2600278592 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 20 |
Hb_039547_010 |
0.2621111303 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |