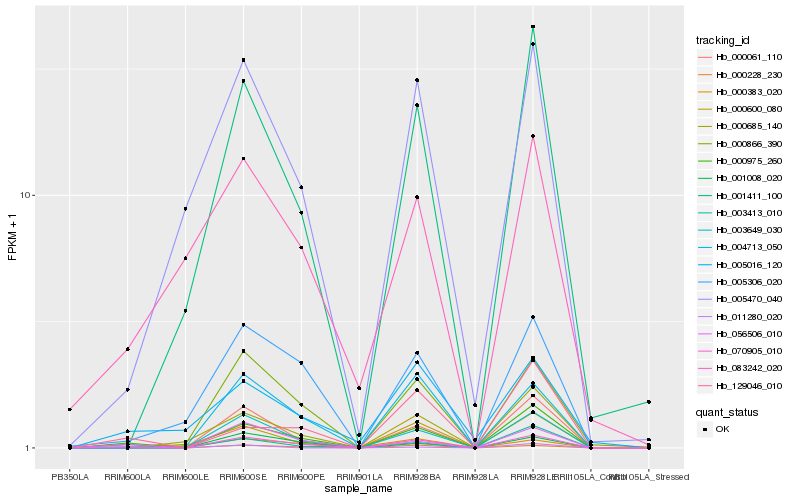
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_070905_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 2 |
Hb_000228_230 |
0.0998423592 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_000975_260 |
0.1228152788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000383_020 |
0.2171280581 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 5 |
Hb_003413_010 |
0.2234175157 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 6 |
Hb_003649_030 |
0.2298230501 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 7 |
Hb_005306_020 |
0.2362060802 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 8 |
Hb_000866_390 |
0.2427315355 |
- |
- |
PREDICTED: O-glucosyltransferase rumi homolog [Jatropha curcas] |
| 9 |
Hb_004713_050 |
0.2450730467 |
- |
- |
RecName: Full=Beta-amyrin synthase [Betula platyphylla] |
| 10 |
Hb_000061_110 |
0.2459379376 |
- |
- |
- |
| 11 |
Hb_005016_120 |
0.2469753733 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 12 |
Hb_129046_010 |
0.2574716336 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 13 |
Hb_001411_100 |
0.2605968395 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 14 |
Hb_005470_040 |
0.2647790672 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 15 |
Hb_001008_020 |
0.2652493045 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000600_080 |
0.2667571578 |
- |
- |
- |
| 17 |
Hb_056506_010 |
0.2678750741 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 18 |
Hb_000685_140 |
0.2697092056 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 19 |
Hb_011280_020 |
0.2713900611 |
- |
- |
polyprotein [Oryza australiensis] |
| 20 |
Hb_083242_020 |
0.2726483066 |
- |
- |
hypothetical protein JCGZ_20097 [Jatropha curcas] |