| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
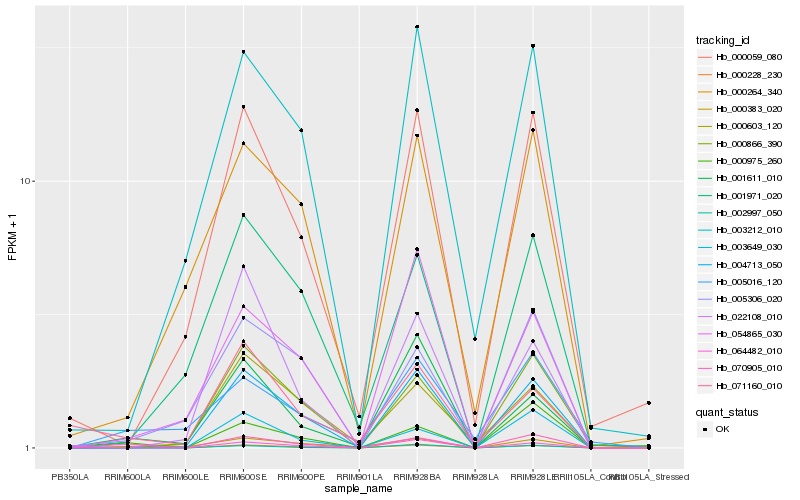
Hb_000228_230 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_070905_010 |
0.0998423592 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 3 |
Hb_000975_260 |
0.1749666101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000383_020 |
0.1777387856 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 5 |
Hb_004713_050 |
0.1902150376 |
- |
- |
RecName: Full=Beta-amyrin synthase [Betula platyphylla] |
| 6 |
Hb_001611_010 |
0.203519093 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 7 |
Hb_054865_030 |
0.2167883701 |
- |
- |
- |
| 8 |
Hb_002997_050 |
0.2179240465 |
- |
- |
PREDICTED: receptor-like protein 12 isoform X4 [Populus euphratica] |
| 9 |
Hb_064482_010 |
0.2208643496 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_000866_390 |
0.2235521006 |
- |
- |
PREDICTED: O-glucosyltransferase rumi homolog [Jatropha curcas] |
| 11 |
Hb_005306_020 |
0.2286104374 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 12 |
Hb_005016_120 |
0.2323030957 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 13 |
Hb_000603_120 |
0.2373895261 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_003649_030 |
0.239164789 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 15 |
Hb_003212_010 |
0.2405627015 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 16 |
Hb_000059_080 |
0.242398674 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 17 |
Hb_071160_010 |
0.2471906351 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 18 |
Hb_000264_340 |
0.2478289161 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_022108_010 |
0.2494871896 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Glycine soja] |
| 20 |
Hb_001971_020 |
0.2494894809 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Populus euphratica] |