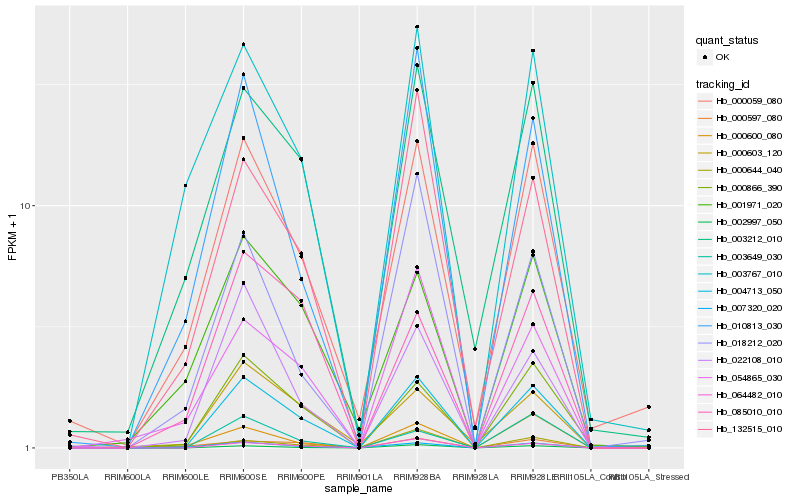
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004713_050 |
0.0 |
- |
- |
RecName: Full=Beta-amyrin synthase [Betula platyphylla] |
| 2 |
Hb_002997_050 |
0.1181902884 |
- |
- |
PREDICTED: receptor-like protein 12 isoform X4 [Populus euphratica] |
| 3 |
Hb_064482_010 |
0.1195809139 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_000866_390 |
0.1197780527 |
- |
- |
PREDICTED: O-glucosyltransferase rumi homolog [Jatropha curcas] |
| 5 |
Hb_000059_080 |
0.1479092347 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 6 |
Hb_022108_010 |
0.1527222397 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Glycine soja] |
| 7 |
Hb_007320_020 |
0.1538659419 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 8 |
Hb_003212_010 |
0.1539843352 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 9 |
Hb_054865_030 |
0.1580348161 |
- |
- |
- |
| 10 |
Hb_010813_030 |
0.1609464547 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 11 |
Hb_132515_010 |
0.163934236 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 12 |
Hb_003649_030 |
0.1662351031 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 13 |
Hb_003767_010 |
0.1706238575 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 14 |
Hb_000603_120 |
0.1713760315 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000644_040 |
0.17167943 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_009157 [Vitis vinifera] |
| 16 |
Hb_000600_080 |
0.1727251095 |
- |
- |
- |
| 17 |
Hb_000597_080 |
0.1756487613 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_001971_020 |
0.1778977307 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Populus euphratica] |
| 19 |
Hb_085010_010 |
0.1825015123 |
- |
- |
PREDICTED: MLO-like protein 3 isoform X2 [Vitis vinifera] |
| 20 |
Hb_018212_020 |
0.1835080235 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |