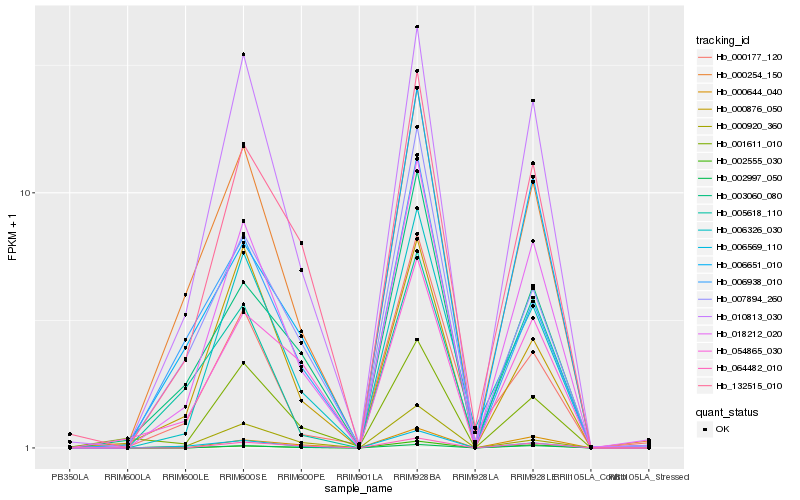
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018212_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 2 |
Hb_010813_030 |
0.0771883789 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 3 |
Hb_000254_150 |
0.0967963894 |
- |
- |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 4 |
Hb_132515_010 |
0.1077164815 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 5 |
Hb_006326_030 |
0.1092940535 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_006569_110 |
0.116759251 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002997_050 |
0.1275947327 |
- |
- |
PREDICTED: receptor-like protein 12 isoform X4 [Populus euphratica] |
| 8 |
Hb_064482_010 |
0.1338111336 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_003060_080 |
0.1377242847 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_000644_040 |
0.1411668494 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_009157 [Vitis vinifera] |
| 11 |
Hb_005618_110 |
0.143331034 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_054865_030 |
0.1434606035 |
- |
- |
- |
| 13 |
Hb_002555_030 |
0.1461128745 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Malus domestica] |
| 14 |
Hb_000876_050 |
0.1495250279 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 15 |
Hb_001611_010 |
0.1507821128 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 16 |
Hb_000920_360 |
0.1536295761 |
- |
- |
PREDICTED: (R,S)-reticuline 7-O-methyltransferase-like [Jatropha curcas] |
| 17 |
Hb_006651_010 |
0.1548761984 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4-like [Citrus sinensis] |
| 18 |
Hb_000177_120 |
0.1559940737 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007894_260 |
0.1575589203 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 20 |
Hb_006938_010 |
0.159050045 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |