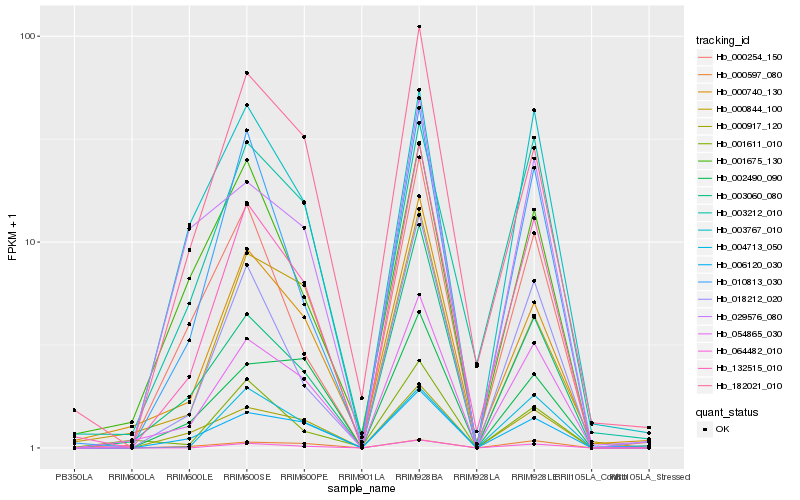
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054865_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_132515_010 |
0.0939929625 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 3 |
Hb_006120_030 |
0.0992592727 |
- |
- |
- |
| 4 |
Hb_000917_120 |
0.1147443465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000740_130 |
0.1220147126 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 6 |
Hb_003060_080 |
0.1246497702 |
- |
- |
PREDICTED: acyl-CoA--sterol O-acyltransferase 1 [Jatropha curcas] |
| 7 |
Hb_064482_010 |
0.1353624894 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_002490_090 |
0.141525584 |
- |
- |
PREDICTED: uncharacterized protein LOC105133003 [Populus euphratica] |
| 9 |
Hb_000844_100 |
0.1417502259 |
- |
- |
hypothetical protein JCGZ_04351 [Jatropha curcas] |
| 10 |
Hb_018212_020 |
0.1434606035 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 11 |
Hb_003212_010 |
0.1441455071 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 12 |
Hb_000597_080 |
0.1463136222 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_003767_010 |
0.1473578733 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 14 |
Hb_001611_010 |
0.1503363916 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 15 |
Hb_029576_080 |
0.1504941111 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 16 |
Hb_010813_030 |
0.1538448055 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 17 |
Hb_182021_010 |
0.153989208 |
- |
- |
sucrose transporter 4 [Hevea brasiliensis] |
| 18 |
Hb_000254_150 |
0.1566533259 |
- |
- |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 19 |
Hb_001675_130 |
0.1577827268 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein GLABRA 2 [Jatropha curcas] |
| 20 |
Hb_004713_050 |
0.1580348161 |
- |
- |
RecName: Full=Beta-amyrin synthase [Betula platyphylla] |