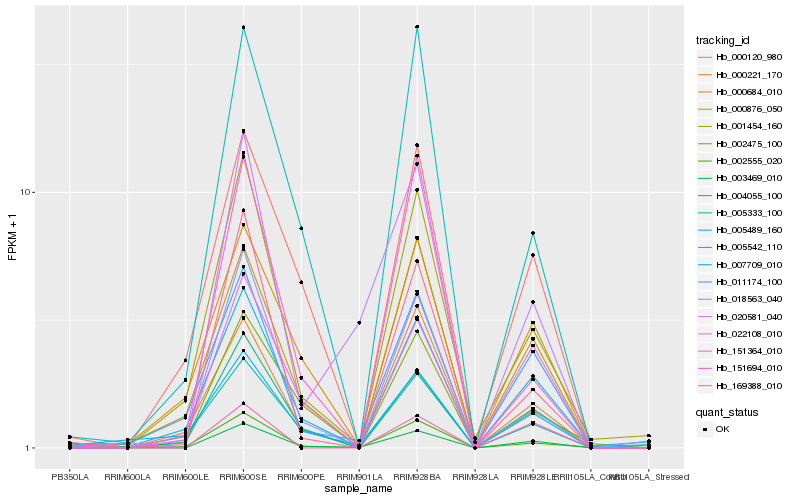
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003469_010 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 2 |
Hb_004055_100 |
0.1317298675 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 3 |
Hb_005542_110 |
0.1477597444 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 4 |
Hb_022108_010 |
0.1486519129 |
- |
- |
G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Glycine soja] |
| 5 |
Hb_020581_040 |
0.1498536045 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like isoform X3 [Sesamum indicum] |
| 6 |
Hb_001454_160 |
0.1600909489 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 7 |
Hb_011174_100 |
0.1602757021 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_169388_010 |
0.166684439 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 9 |
Hb_000876_050 |
0.1674736373 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 10 |
Hb_018563_040 |
0.1690840789 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_002555_020 |
0.1704772275 |
- |
- |
Receptor-like protein kinase [Medicago truncatula] |
| 12 |
Hb_005489_160 |
0.1706945285 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 13 |
Hb_005333_100 |
0.1826775099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000221_170 |
0.183298103 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 2 [Jatropha curcas] |
| 15 |
Hb_151694_010 |
0.1835597877 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000120_980 |
0.1841663584 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 17 |
Hb_000684_010 |
0.1847639399 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_002475_100 |
0.1877162928 |
- |
- |
PREDICTED: uncharacterized protein LOC105645451 [Jatropha curcas] |
| 19 |
Hb_007709_010 |
0.1939692622 |
- |
- |
- |
| 20 |
Hb_151364_010 |
0.1948008072 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |