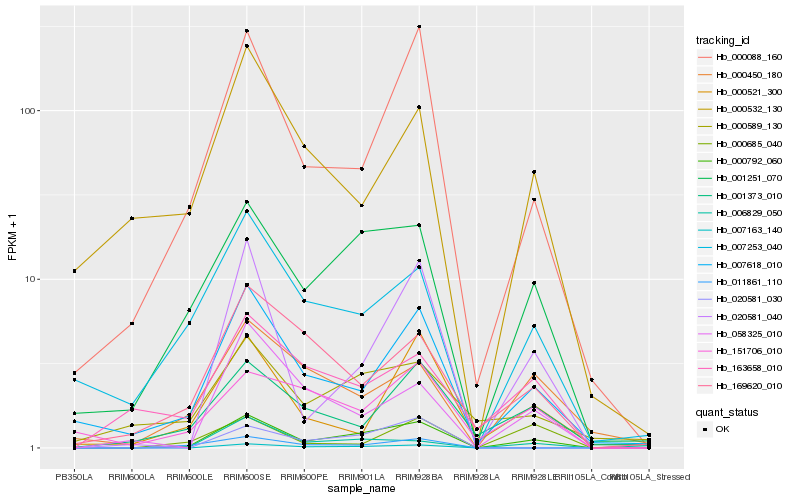
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000792_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001251_070 |
0.1694972939 |
- |
- |
PREDICTED: probable aldo-keto reductase 2, partial [Populus euphratica] |
| 3 |
Hb_000088_160 |
0.1922866546 |
- |
- |
PREDICTED: uncharacterized protein LOC105629775 [Jatropha curcas] |
| 4 |
Hb_007618_010 |
0.1991885495 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 5 |
Hb_020581_040 |
0.2082223245 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like isoform X3 [Sesamum indicum] |
| 6 |
Hb_007163_140 |
0.2351854797 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_12606 [Jatropha curcas] |
| 7 |
Hb_007253_040 |
0.2391110529 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001373_010 |
0.2446097828 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 9 |
Hb_058325_010 |
0.2496989504 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 10 |
Hb_006829_050 |
0.2501053692 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 11 |
Hb_169620_010 |
0.2515367553 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 12 |
Hb_000450_180 |
0.2539165042 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 13 |
Hb_000521_300 |
0.2545823888 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 14 |
Hb_163658_010 |
0.2551091446 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 15 |
Hb_151706_010 |
0.2555456853 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 16 |
Hb_020581_030 |
0.2556725431 |
- |
- |
- |
| 17 |
Hb_011861_110 |
0.258466906 |
- |
- |
PREDICTED: uncharacterized protein LOC105636580 [Jatropha curcas] |
| 18 |
Hb_000589_130 |
0.2588803944 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_000532_130 |
0.2643536162 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |
| 20 |
Hb_000685_040 |
0.2651481548 |
- |
- |
hypothetical protein OsJ_17676 [Oryza sativa Japonica Group] |