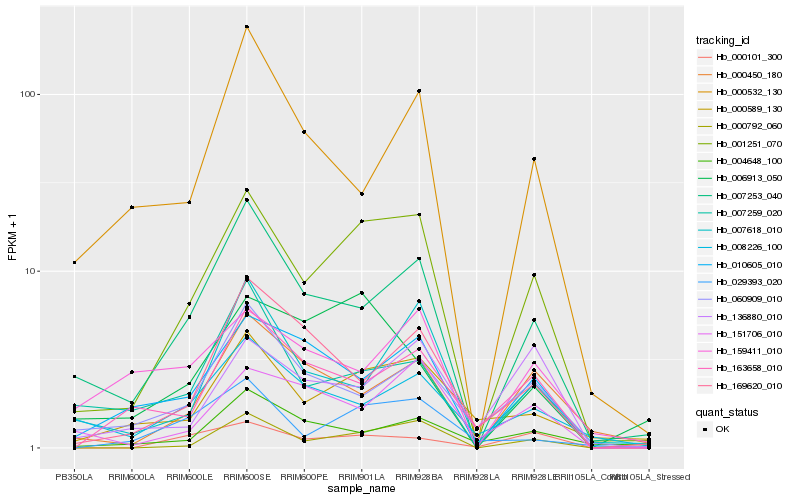
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001251_070 |
0.0 |
- |
- |
PREDICTED: probable aldo-keto reductase 2, partial [Populus euphratica] |
| 2 |
Hb_000792_060 |
0.1694972939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007253_040 |
0.1787404062 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000589_130 |
0.2052928426 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_010605_010 |
0.2086293799 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 6 |
Hb_000450_180 |
0.2140493214 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 7 |
Hb_029393_020 |
0.2214807081 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 12-like [Jatropha curcas] |
| 8 |
Hb_007618_010 |
0.2258615459 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 9 |
Hb_000101_300 |
0.2269346085 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein B456_002G046500, partial [Gossypium raimondii] |
| 10 |
Hb_151706_010 |
0.2278389719 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 11 |
Hb_060909_010 |
0.2280343905 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 12 |
Hb_163658_010 |
0.2301851137 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 13 |
Hb_007259_020 |
0.231733852 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 14 |
Hb_136880_010 |
0.236295589 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 15 |
Hb_008226_100 |
0.2403382763 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 16 |
Hb_006913_050 |
0.2407303213 |
- |
- |
- |
| 17 |
Hb_169620_010 |
0.2423821833 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 18 |
Hb_000532_130 |
0.2478471448 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |
| 19 |
Hb_004648_100 |
0.2482566096 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 20 |
Hb_159411_010 |
0.2514326996 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |