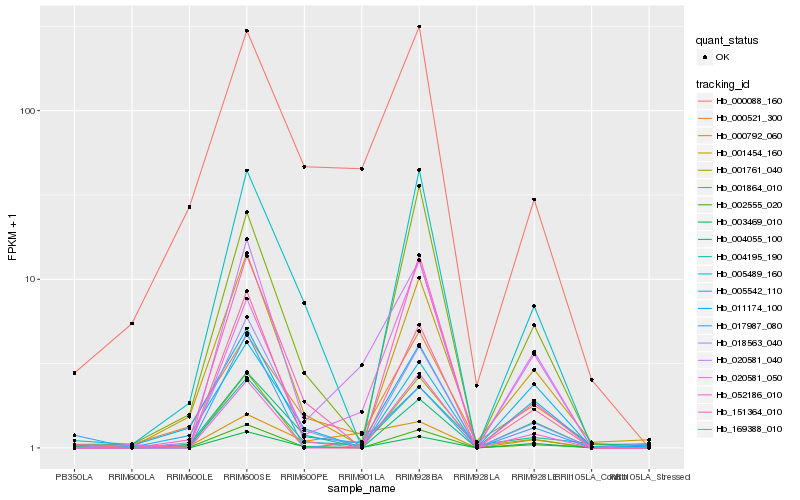
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020581_040 |
0.0 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075-like isoform X3 [Sesamum indicum] |
| 2 |
Hb_003469_010 |
0.1498536045 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 3 |
Hb_020581_050 |
0.1915722651 |
- |
- |
- |
| 4 |
Hb_002555_020 |
0.2019892506 |
- |
- |
Receptor-like protein kinase [Medicago truncatula] |
| 5 |
Hb_169388_010 |
0.2033747378 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 6 |
Hb_000792_060 |
0.2082223245 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000088_160 |
0.2100420288 |
- |
- |
PREDICTED: uncharacterized protein LOC105629775 [Jatropha curcas] |
| 8 |
Hb_001454_160 |
0.2145088219 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 9 |
Hb_011174_100 |
0.2175877309 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_004195_190 |
0.2184956573 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_018563_040 |
0.219834014 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_052186_010 |
0.2236275524 |
- |
- |
hypothetical protein JCGZ_17055 [Jatropha curcas] |
| 13 |
Hb_151364_010 |
0.2267015401 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 14 |
Hb_000521_300 |
0.2305059146 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 15 |
Hb_001761_040 |
0.2316069347 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Jatropha curcas] |
| 16 |
Hb_001864_010 |
0.2316466909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005489_160 |
0.2329459098 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 18 |
Hb_017987_080 |
0.2366735616 |
- |
- |
Uncharacterized protein TCM_001832 [Theobroma cacao] |
| 19 |
Hb_005542_110 |
0.2374988321 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 20 |
Hb_004055_100 |
0.2388019552 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |