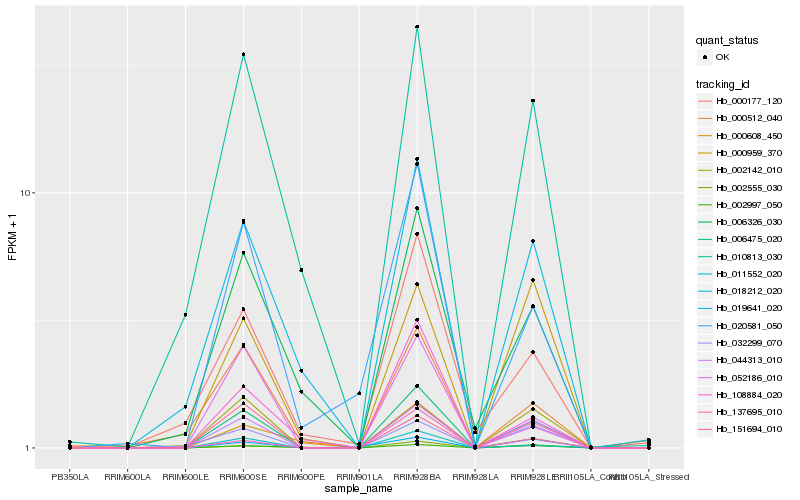
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006475_020 |
0.0 |
- |
- |
PREDICTED: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |
| 2 |
Hb_044313_010 |
0.0751789162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_011552_020 |
0.1289843563 |
- |
- |
hypothetical protein F383_28301 [Gossypium arboreum] |
| 4 |
Hb_020581_050 |
0.1644667786 |
- |
- |
- |
| 5 |
Hb_002142_010 |
0.1676491486 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_032299_070 |
0.169942646 |
- |
- |
- |
| 7 |
Hb_018212_020 |
0.1746899477 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 8 |
Hb_108884_020 |
0.1803159492 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 9 |
Hb_006326_030 |
0.1811414817 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_151694_010 |
0.1887038127 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_010813_030 |
0.1897919433 |
- |
- |
PREDICTED: indole-3-acetaldehyde oxidase-like [Jatropha curcas] |
| 12 |
Hb_000177_120 |
0.1907769408 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002997_050 |
0.1931111153 |
- |
- |
PREDICTED: receptor-like protein 12 isoform X4 [Populus euphratica] |
| 14 |
Hb_002555_030 |
0.1939267084 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Malus domestica] |
| 15 |
Hb_052186_010 |
0.1950727323 |
- |
- |
hypothetical protein JCGZ_17055 [Jatropha curcas] |
| 16 |
Hb_137695_010 |
0.1969395664 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like [Malus domestica] |
| 17 |
Hb_000959_370 |
0.2008012304 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 18 |
Hb_019641_020 |
0.2018050016 |
- |
- |
hypothetical protein AALP_AAs49670U000100, partial [Arabis alpina] |
| 19 |
Hb_000512_040 |
0.2121070291 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 20 |
Hb_000608_450 |
0.2147474581 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |