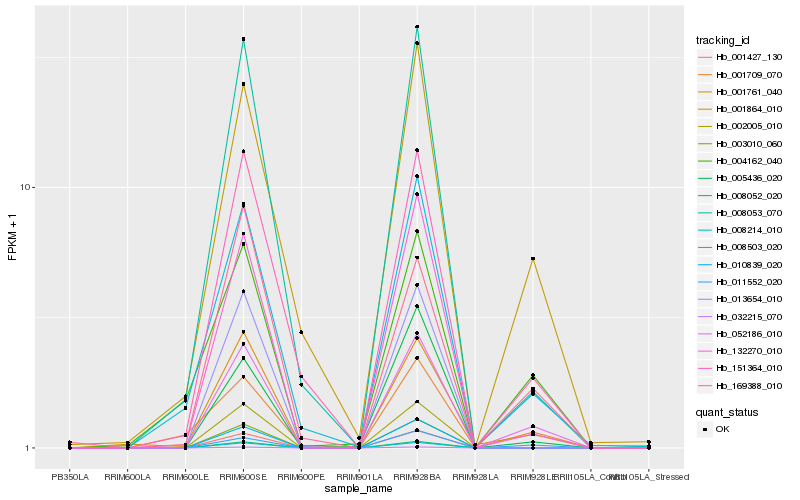
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052186_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_17055 [Jatropha curcas] |
| 2 |
Hb_011552_020 |
0.0954529332 |
- |
- |
hypothetical protein F383_28301 [Gossypium arboreum] |
| 3 |
Hb_001761_040 |
0.1271365526 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Jatropha curcas] |
| 4 |
Hb_132270_010 |
0.1386112202 |
- |
- |
hypothetical protein JCGZ_25438 [Jatropha curcas] |
| 5 |
Hb_008503_020 |
0.1453820601 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 6 |
Hb_001864_010 |
0.1499012139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_151364_010 |
0.149905567 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 8 |
Hb_008053_070 |
0.1575556371 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X4 [Jatropha curcas] |
| 9 |
Hb_004162_040 |
0.1586205891 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase-like [Crassostrea gigas] |
| 10 |
Hb_005436_020 |
0.1615207674 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_169388_010 |
0.1633569457 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 12 |
Hb_032215_070 |
0.1759316408 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_001427_130 |
0.1759621311 |
- |
- |
PREDICTED: uncharacterized protein LOC101510272 [Cicer arietinum] |
| 14 |
Hb_003010_060 |
0.1763257199 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 15 |
Hb_008052_020 |
0.1765626568 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 16 |
Hb_008214_010 |
0.1766062986 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 17 |
Hb_002005_010 |
0.1770880946 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB39-like isoform X2 [Populus euphratica] |
| 18 |
Hb_013654_010 |
0.179270661 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_001709_070 |
0.1794869044 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 20 |
Hb_010839_020 |
0.1797850837 |
- |
- |
- |