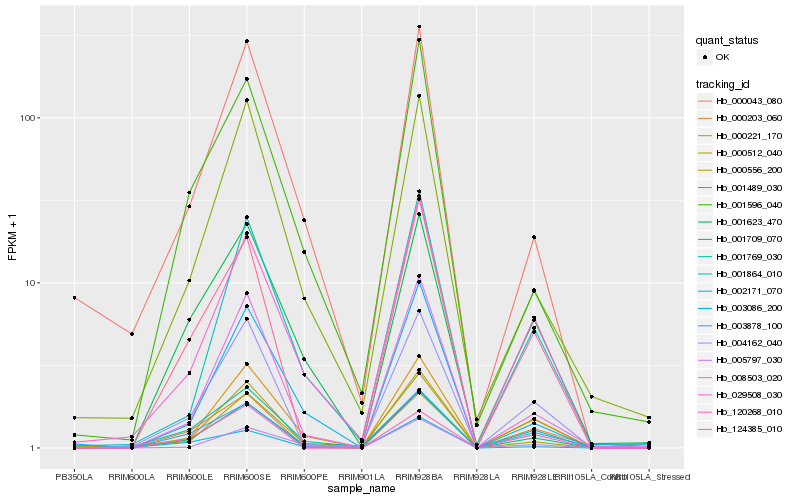
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001709_070 |
0.0 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 2 |
Hb_004162_040 |
0.0835420745 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase-like [Crassostrea gigas] |
| 3 |
Hb_124385_010 |
0.0861221573 |
- |
- |
hypothetical protein JCGZ_13882 [Jatropha curcas] |
| 4 |
Hb_008503_020 |
0.0988429 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 5 |
Hb_000221_170 |
0.1060829774 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 2 [Jatropha curcas] |
| 6 |
Hb_029508_030 |
0.1081735073 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0002s04880g [Populus trichocarpa] |
| 7 |
Hb_001623_470 |
0.1274636232 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 8 |
Hb_001864_010 |
0.1313320406 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001489_030 |
0.1356688525 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 10 |
Hb_000556_200 |
0.1359003336 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 22 [Jatropha curcas] |
| 11 |
Hb_003086_200 |
0.1373606984 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 12 |
Hb_000203_060 |
0.1392246563 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0004s05660g [Populus trichocarpa] |
| 13 |
Hb_001596_040 |
0.140468229 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase 1-like [Populus euphratica] |
| 14 |
Hb_000043_080 |
0.1426205702 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 15 |
Hb_000512_040 |
0.1452837188 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 16 |
Hb_001769_030 |
0.1465392468 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 17 |
Hb_003878_100 |
0.147531648 |
desease resistance |
Gene Name: NB-ARC |
LRR and NB-ARC domains-containing disease resistance protein, putative [Theobroma cacao] |
| 18 |
Hb_120268_010 |
0.1481964736 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1-like isoform X3 [Populus euphratica] |
| 19 |
Hb_005797_030 |
0.1485118114 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like [Jatropha curcas] |
| 20 |
Hb_002171_070 |
0.1499075649 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance gene-like protein ARGH30 [Populus trichocarpa] |