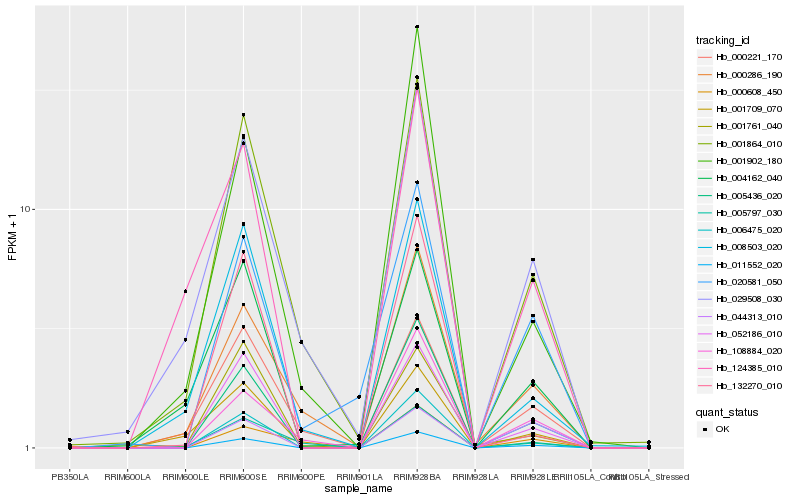
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011552_020 |
0.0 |
- |
- |
hypothetical protein F383_28301 [Gossypium arboreum] |
| 2 |
Hb_052186_010 |
0.0954529332 |
- |
- |
hypothetical protein JCGZ_17055 [Jatropha curcas] |
| 3 |
Hb_006475_020 |
0.1289843563 |
- |
- |
PREDICTED: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |
| 4 |
Hb_108884_020 |
0.1458918693 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 5 |
Hb_020581_050 |
0.1485542708 |
- |
- |
- |
| 6 |
Hb_001864_010 |
0.1512455904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005436_020 |
0.1583290165 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_132270_010 |
0.1745739797 |
- |
- |
hypothetical protein JCGZ_25438 [Jatropha curcas] |
| 9 |
Hb_001902_180 |
0.1753551717 |
- |
- |
PREDICTED: uncharacterized protein LOC105129658 [Populus euphratica] |
| 10 |
Hb_000286_190 |
0.1762071616 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 11 |
Hb_008503_020 |
0.1764701379 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 12 |
Hb_124385_010 |
0.1795061319 |
- |
- |
hypothetical protein JCGZ_13882 [Jatropha curcas] |
| 13 |
Hb_004162_040 |
0.1796413695 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase-like [Crassostrea gigas] |
| 14 |
Hb_029508_030 |
0.1838535211 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0002s04880g [Populus trichocarpa] |
| 15 |
Hb_001709_070 |
0.1855759171 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 16 |
Hb_005797_030 |
0.1881738135 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like [Jatropha curcas] |
| 17 |
Hb_000608_450 |
0.192432571 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105645015 [Jatropha curcas] |
| 18 |
Hb_000221_170 |
0.1944720046 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 2 [Jatropha curcas] |
| 19 |
Hb_001761_040 |
0.194803761 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Jatropha curcas] |
| 20 |
Hb_044313_010 |
0.1956386552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |