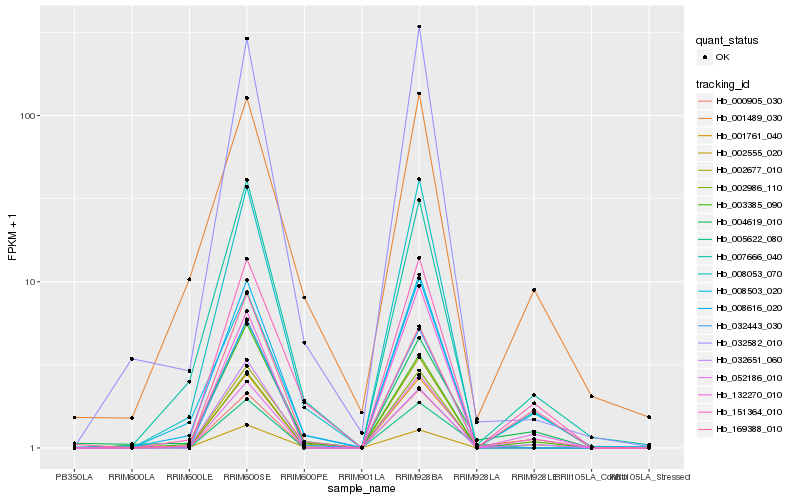
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001761_040 |
0.0 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Jatropha curcas] |
| 2 |
Hb_004619_010 |
0.0964831833 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 3 |
Hb_052186_010 |
0.1271365526 |
- |
- |
hypothetical protein JCGZ_17055 [Jatropha curcas] |
| 4 |
Hb_008053_070 |
0.1387946722 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X4 [Jatropha curcas] |
| 5 |
Hb_007666_040 |
0.1429291543 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_032651_060 |
0.1438359317 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_151364_010 |
0.1445582092 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 8 |
Hb_169388_010 |
0.1472974737 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 9 |
Hb_032582_010 |
0.1499092147 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 10 |
Hb_008503_020 |
0.1513732483 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 11 |
Hb_003385_090 |
0.155252751 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 12 |
Hb_005622_080 |
0.1571770303 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 13 |
Hb_132270_010 |
0.160299738 |
- |
- |
hypothetical protein JCGZ_25438 [Jatropha curcas] |
| 14 |
Hb_002677_010 |
0.1657481753 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2 [Populus euphratica] |
| 15 |
Hb_001489_030 |
0.166405592 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 16 |
Hb_002555_020 |
0.1678389506 |
- |
- |
Receptor-like protein kinase [Medicago truncatula] |
| 17 |
Hb_000905_030 |
0.1689320398 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 18 |
Hb_008616_020 |
0.169158791 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_032443_030 |
0.1691630424 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 20 |
Hb_002986_110 |
0.1699892403 |
- |
- |
PREDICTED: patatin-like protein 1 [Jatropha curcas] |