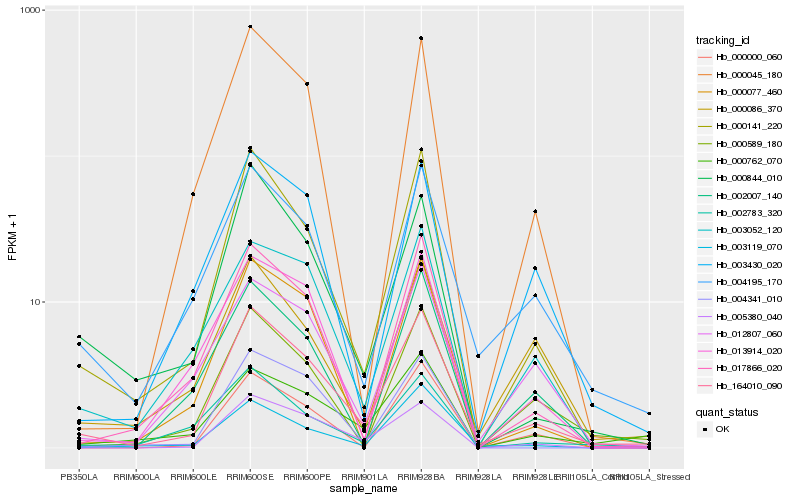
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164010_090 |
0.0 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 2 |
Hb_000141_220 |
0.0976780528 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Gossypium raimondii] |
| 3 |
Hb_004195_170 |
0.1291452776 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_005380_040 |
0.1359707663 |
- |
- |
hypothetical protein PRUPE_ppa015877mg, partial [Prunus persica] |
| 5 |
Hb_017866_020 |
0.1420399037 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 40 [Jatropha curcas] |
| 6 |
Hb_000589_180 |
0.1454901231 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 9 [Jatropha curcas] |
| 7 |
Hb_000844_010 |
0.1485279122 |
- |
- |
PREDICTED: lysine histidine transporter-like 8 [Jatropha curcas] |
| 8 |
Hb_000045_180 |
0.1497031412 |
- |
- |
aquaporin PIP1 [Hevea brasiliensis] |
| 9 |
Hb_003052_120 |
0.1499260212 |
- |
- |
hypothetical protein POPTR_0008s10970g [Populus trichocarpa] |
| 10 |
Hb_000077_460 |
0.1529927774 |
- |
- |
- |
| 11 |
Hb_003119_070 |
0.1530467448 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 12 |
Hb_012807_060 |
0.1555324127 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_002783_320 |
0.1560151543 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FC isoform X1 [Jatropha curcas] |
| 14 |
Hb_003430_020 |
0.1577951534 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_002007_140 |
0.1610321525 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like [Populus euphratica] |
| 16 |
Hb_000000_060 |
0.1610524418 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 17 |
Hb_000762_070 |
0.1629645042 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 18 |
Hb_000086_370 |
0.1630325498 |
- |
- |
Long-chain-alcohol oxidase FAO2 [Triticum urartu] |
| 19 |
Hb_004341_010 |
0.1641425896 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 20 |
Hb_013914_020 |
0.1646537596 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |