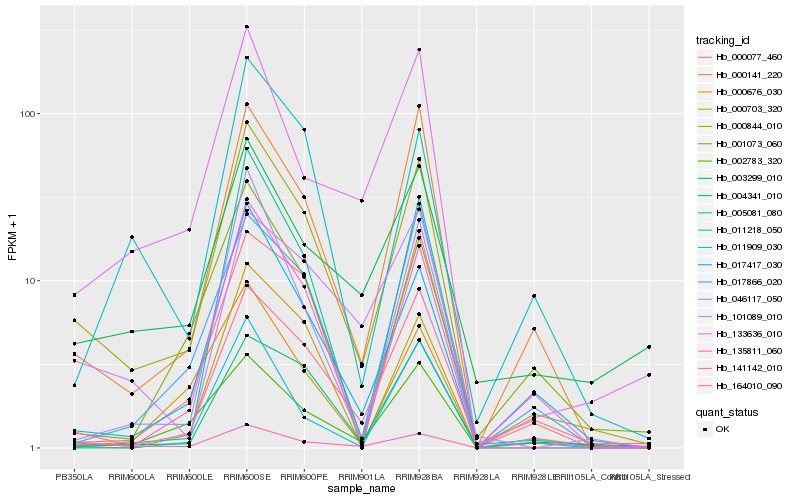
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000844_010 |
0.0 |
- |
- |
PREDICTED: lysine histidine transporter-like 8 [Jatropha curcas] |
| 2 |
Hb_000141_220 |
0.1362956699 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Gossypium raimondii] |
| 3 |
Hb_017417_030 |
0.138689462 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 4 |
Hb_164010_090 |
0.1485279122 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 5 |
Hb_141142_010 |
0.1523169894 |
- |
- |
hypothetical protein JCGZ_21320 [Jatropha curcas] |
| 6 |
Hb_135811_060 |
0.1562111027 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 7 |
Hb_133636_010 |
0.1640507388 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 8 |
Hb_002783_320 |
0.1651187272 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FC isoform X1 [Jatropha curcas] |
| 9 |
Hb_046117_050 |
0.1676218114 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0009s14590g [Populus trichocarpa] |
| 10 |
Hb_004341_010 |
0.1686087931 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 11 |
Hb_000703_320 |
0.1770899312 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 12 |
Hb_003299_010 |
0.1783599832 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 13 |
Hb_000676_030 |
0.179265407 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-B [Jatropha curcas] |
| 14 |
Hb_011218_050 |
0.1823302076 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 15 |
Hb_000077_460 |
0.1825368335 |
- |
- |
- |
| 16 |
Hb_017866_020 |
0.1830615095 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 40 [Jatropha curcas] |
| 17 |
Hb_101089_010 |
0.1836449759 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 18 |
Hb_011909_030 |
0.1849629805 |
- |
- |
hypothetical protein JCGZ_00732 [Jatropha curcas] |
| 19 |
Hb_001073_060 |
0.185220726 |
- |
- |
phosphate transporter 1 [Hevea brasiliensis] |
| 20 |
Hb_005081_080 |
0.1865405888 |
- |
- |
hypothetical protein JCGZ_16787 [Jatropha curcas] |