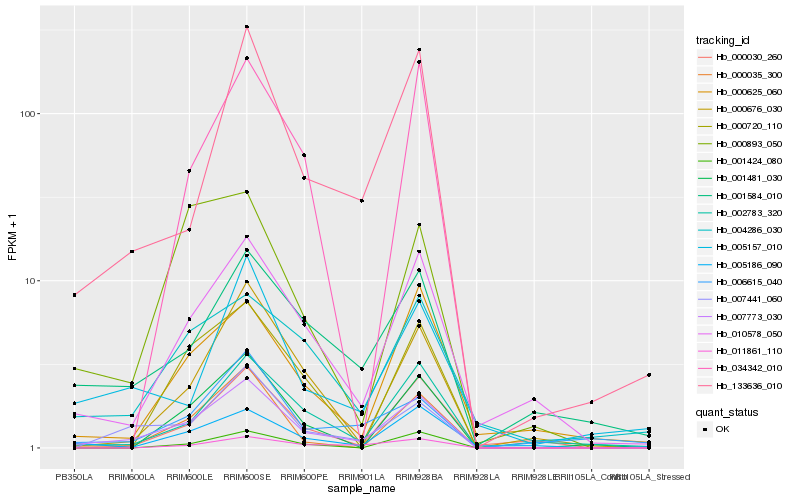
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007773_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_007441_060 |
0.1523562282 |
- |
- |
- |
| 3 |
Hb_010578_050 |
0.1651469809 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_133636_010 |
0.1733290224 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 5 |
Hb_000035_300 |
0.1803208235 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002783_320 |
0.1833951008 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FC isoform X1 [Jatropha curcas] |
| 7 |
Hb_001584_010 |
0.1894335132 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 8 |
Hb_000676_030 |
0.1922995177 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-B [Jatropha curcas] |
| 9 |
Hb_005186_090 |
0.2010935532 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_004286_030 |
0.2055881197 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 11 |
Hb_000625_060 |
0.2060513696 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 12 |
Hb_001481_030 |
0.2061064518 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_000030_260 |
0.2079979532 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 14 |
Hb_001424_080 |
0.2127488129 |
- |
- |
PREDICTED: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |
| 15 |
Hb_011861_110 |
0.2132311665 |
- |
- |
PREDICTED: uncharacterized protein LOC105636580 [Jatropha curcas] |
| 16 |
Hb_000893_050 |
0.2144949521 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 17 |
Hb_000720_110 |
0.2162758559 |
- |
- |
hypothetical protein JCGZ_20628 [Jatropha curcas] |
| 18 |
Hb_005157_010 |
0.2167288389 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 19 |
Hb_006615_040 |
0.2174186143 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 20 |
Hb_034342_010 |
0.2200740156 |
- |
- |
conserved hypothetical protein [Ricinus communis] |