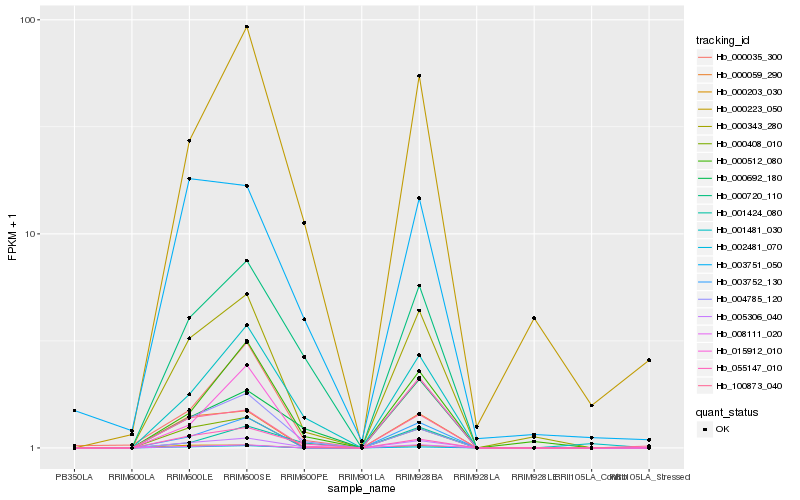
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005306_040 |
0.0 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 2 |
Hb_000343_280 |
0.0954159302 |
- |
- |
hypothetical protein JCGZ_24094 [Jatropha curcas] |
| 3 |
Hb_000720_110 |
0.1120801719 |
- |
- |
hypothetical protein JCGZ_20628 [Jatropha curcas] |
| 4 |
Hb_001481_030 |
0.1274407398 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_055147_010 |
0.1375527676 |
- |
- |
thioredoxin H-type 7 [Hevea brasiliensis] |
| 6 |
Hb_100873_040 |
0.1438601181 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000692_180 |
0.1516396416 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 8 |
Hb_000408_010 |
0.151708474 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 9 |
Hb_000035_300 |
0.1545882115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000223_050 |
0.1586536451 |
- |
- |
PREDICTED: endochitinase A-like [Jatropha curcas] |
| 11 |
Hb_000059_290 |
0.1587229768 |
- |
- |
pterin-4-alpha-carbinolamine dehydratase, putative [Ricinus communis] |
| 12 |
Hb_008111_020 |
0.158793789 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 13 |
Hb_004785_120 |
0.1599319808 |
- |
- |
Receptor protein kinase 1 [Theobroma cacao] |
| 14 |
Hb_003752_130 |
0.164249656 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 15 |
Hb_000512_080 |
0.1642670035 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_002481_070 |
0.1673773911 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 17 |
Hb_003751_050 |
0.1721213655 |
- |
- |
PREDICTED: amino acid permease 4-like [Jatropha curcas] |
| 18 |
Hb_000203_030 |
0.172479646 |
- |
- |
- |
| 19 |
Hb_001424_080 |
0.1766245842 |
- |
- |
PREDICTED: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |
| 20 |
Hb_015912_010 |
0.181186126 |
- |
- |
cytochrome P450, putative [Ricinus communis] |