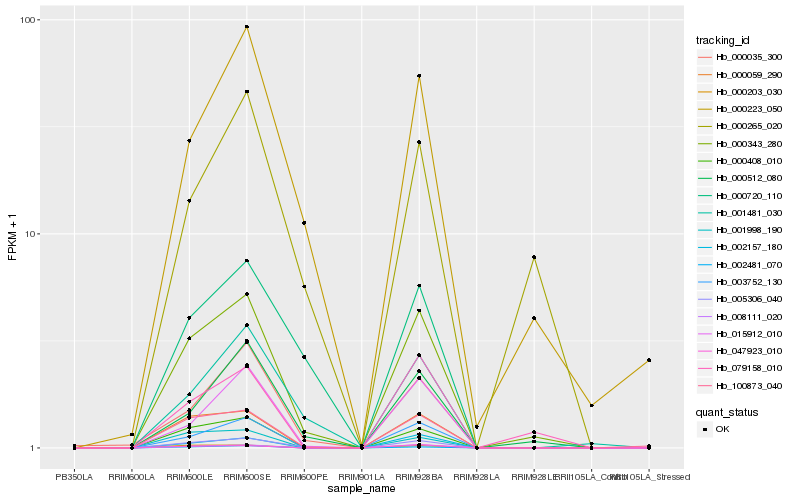
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000343_280 |
0.0 |
- |
- |
hypothetical protein JCGZ_24094 [Jatropha curcas] |
| 2 |
Hb_005306_040 |
0.0954159302 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 3 |
Hb_008111_020 |
0.1388389405 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 4 |
Hb_100873_040 |
0.1409863861 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000512_080 |
0.141528693 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_000059_290 |
0.1457000263 |
- |
- |
pterin-4-alpha-carbinolamine dehydratase, putative [Ricinus communis] |
| 7 |
Hb_000408_010 |
0.1457717919 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 8 |
Hb_003752_130 |
0.1486130659 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 9 |
Hb_000223_050 |
0.154589488 |
- |
- |
PREDICTED: endochitinase A-like [Jatropha curcas] |
| 10 |
Hb_000203_030 |
0.1622187113 |
- |
- |
- |
| 11 |
Hb_079158_010 |
0.1635654078 |
- |
- |
PREDICTED: sperm acrosomal protein FSA-ACR.1-like [Jatropha curcas] |
| 12 |
Hb_002481_070 |
0.1662781655 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 13 |
Hb_047923_010 |
0.1663673041 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 14 |
Hb_000035_300 |
0.1726633609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002157_180 |
0.1745854258 |
- |
- |
separase, putative [Ricinus communis] |
| 16 |
Hb_015912_010 |
0.1750252493 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_001481_030 |
0.1756736652 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_000720_110 |
0.177549642 |
- |
- |
hypothetical protein JCGZ_20628 [Jatropha curcas] |
| 19 |
Hb_000265_020 |
0.1784400901 |
- |
- |
hypothetical protein POPTR_0011s16980g, partial [Populus trichocarpa] |
| 20 |
Hb_001998_190 |
0.1820293817 |
transcription factor |
TF Family: CSD |
PREDICTED: uncharacterized protein LOC105636942 [Jatropha curcas] |