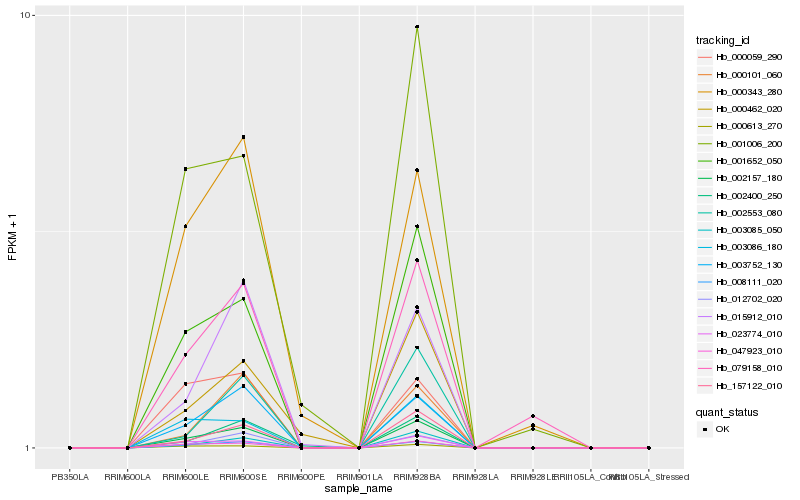
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002157_180 |
0.0 |
- |
- |
separase, putative [Ricinus communis] |
| 2 |
Hb_008111_020 |
0.0623104751 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 3 |
Hb_001652_050 |
0.0729126356 |
- |
- |
- |
| 4 |
Hb_047923_010 |
0.0896654713 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 5 |
Hb_157122_010 |
0.0924020247 |
- |
- |
hypothetical protein JCGZ_21614 [Jatropha curcas] |
| 6 |
Hb_003752_130 |
0.09955764 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 7 |
Hb_003085_050 |
0.1014468345 |
- |
- |
Polygalacturonase non-catalytic subunit AroGP3 precursor, putative [Ricinus communis] |
| 8 |
Hb_023774_010 |
0.1026717115 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 9 |
Hb_000613_270 |
0.1032650783 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 10 |
Hb_012702_020 |
0.1186065062 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 11 |
Hb_003086_180 |
0.128876166 |
- |
- |
PREDICTED: RING-H2 finger protein ATL70-like [Pyrus x bretschneideri] |
| 12 |
Hb_015912_010 |
0.1453828776 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_002553_080 |
0.1556606145 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF096-like [Eucalyptus grandis] |
| 14 |
Hb_001006_200 |
0.1560172121 |
transcription factor |
TF Family: G2-like |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_079158_010 |
0.1563733493 |
- |
- |
PREDICTED: sperm acrosomal protein FSA-ACR.1-like [Jatropha curcas] |
| 16 |
Hb_000101_060 |
0.1570878732 |
- |
- |
- |
| 17 |
Hb_002400_250 |
0.1577769835 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000462_020 |
0.159664704 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000059_290 |
0.1615177154 |
- |
- |
pterin-4-alpha-carbinolamine dehydratase, putative [Ricinus communis] |
| 20 |
Hb_000343_280 |
0.1745854258 |
- |
- |
hypothetical protein JCGZ_24094 [Jatropha curcas] |