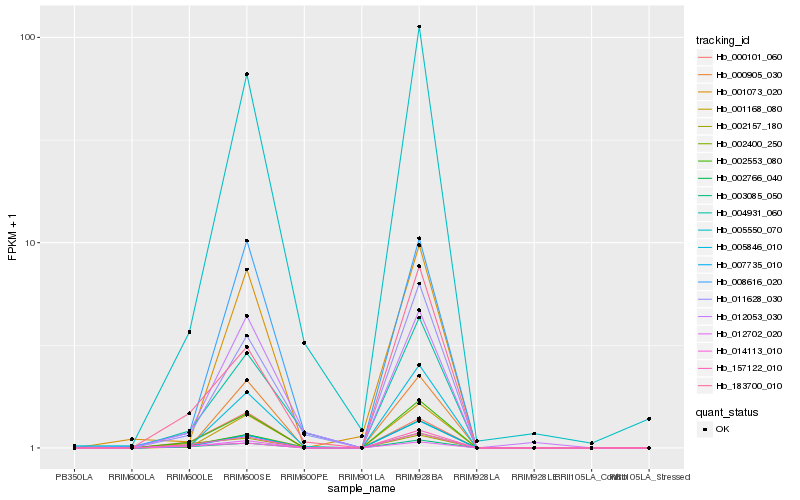
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002553_080 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF096-like [Eucalyptus grandis] |
| 2 |
Hb_003085_050 |
0.0675761626 |
- |
- |
Polygalacturonase non-catalytic subunit AroGP3 precursor, putative [Ricinus communis] |
| 3 |
Hb_157122_010 |
0.0749629152 |
- |
- |
hypothetical protein JCGZ_21614 [Jatropha curcas] |
| 4 |
Hb_002766_040 |
0.0790414795 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 5 |
Hb_007735_010 |
0.0944282368 |
- |
- |
Gibberellin 20 oxidase, putative [Ricinus communis] |
| 6 |
Hb_001168_080 |
0.1198544165 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_005550_070 |
0.1316253574 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Populus euphratica] |
| 8 |
Hb_000101_060 |
0.1341222043 |
- |
- |
- |
| 9 |
Hb_004931_060 |
0.1352000448 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein Myb4 [Nelumbo nucifera] |
| 10 |
Hb_005846_010 |
0.1369680351 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 11 |
Hb_011628_030 |
0.141683906 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002400_250 |
0.147191952 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000905_030 |
0.1477720665 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 14 |
Hb_012702_020 |
0.1479648303 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 15 |
Hb_008616_020 |
0.1524704799 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_183700_010 |
0.1542197257 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_002157_180 |
0.1556606145 |
- |
- |
separase, putative [Ricinus communis] |
| 18 |
Hb_001073_020 |
0.1575424111 |
- |
- |
hypothetical protein CISIN_1g031854mg [Citrus sinensis] |
| 19 |
Hb_012053_030 |
0.1578133064 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g22810 [Jatropha curcas] |
| 20 |
Hb_014113_010 |
0.1580944211 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |