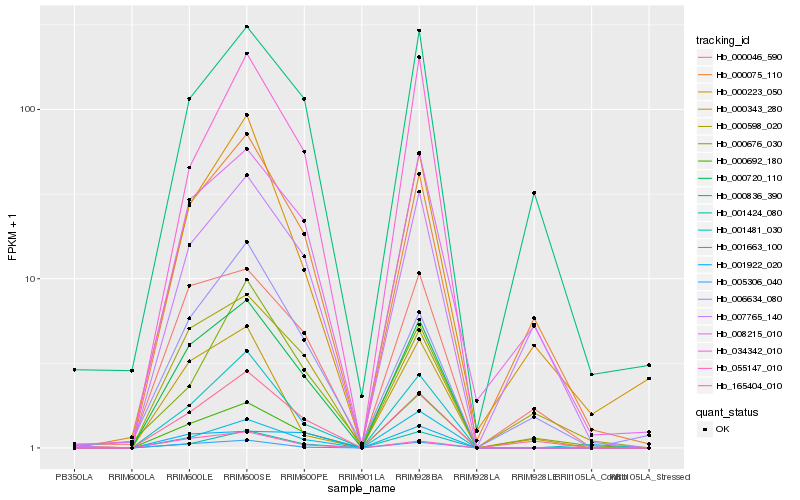
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000720_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_20628 [Jatropha curcas] |
| 2 |
Hb_001481_030 |
0.1118600789 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_005306_040 |
0.1120801719 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 4 |
Hb_000692_180 |
0.1128055341 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 5 |
Hb_165404_010 |
0.1222631805 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 6 |
Hb_055147_010 |
0.1245455648 |
- |
- |
thioredoxin H-type 7 [Hevea brasiliensis] |
| 7 |
Hb_000075_110 |
0.1377723647 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 8 |
Hb_000046_590 |
0.1383618563 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 9 |
Hb_001424_080 |
0.1391973738 |
- |
- |
PREDICTED: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |
| 10 |
Hb_034342_010 |
0.1403351231 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_008215_010 |
0.1448327408 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001663_100 |
0.1576591032 |
- |
- |
- |
| 13 |
Hb_007765_140 |
0.1584622372 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |
| 14 |
Hb_000223_050 |
0.1620984382 |
- |
- |
PREDICTED: endochitinase A-like [Jatropha curcas] |
| 15 |
Hb_000598_020 |
0.1629086631 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 16 |
Hb_000836_390 |
0.1744883143 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 17 |
Hb_000343_280 |
0.177549642 |
- |
- |
hypothetical protein JCGZ_24094 [Jatropha curcas] |
| 18 |
Hb_006634_080 |
0.1776595234 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 19 |
Hb_001922_020 |
0.1777560976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000676_030 |
0.1855687642 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-B [Jatropha curcas] |