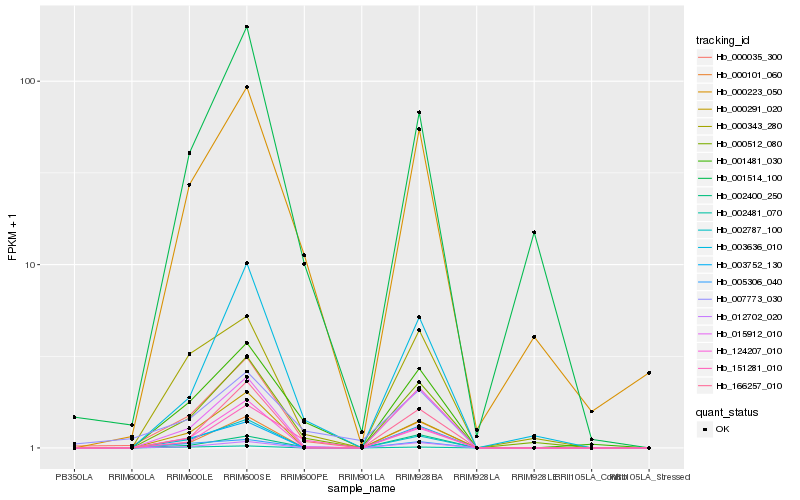
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_300 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000512_080 |
0.1326206807 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_015912_010 |
0.1335645996 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_000291_020 |
0.1461993532 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 5 |
Hb_001481_030 |
0.1479907461 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_002787_100 |
0.1520983789 |
- |
- |
PREDICTED: uncharacterized protein LOC105637209 [Jatropha curcas] |
| 7 |
Hb_005306_040 |
0.1545882115 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 8 |
Hb_003636_010 |
0.1559126835 |
- |
- |
hypothetical protein CISIN_1g037850mg, partial [Citrus sinensis] |
| 9 |
Hb_003752_130 |
0.1588877462 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 10 |
Hb_124207_010 |
0.1612664633 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000223_050 |
0.1634398825 |
- |
- |
PREDICTED: endochitinase A-like [Jatropha curcas] |
| 12 |
Hb_012702_020 |
0.1645667357 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 13 |
Hb_166257_010 |
0.1691291551 |
- |
- |
hypothetical protein JCGZ_21808 [Jatropha curcas] |
| 14 |
Hb_000343_280 |
0.1726633609 |
- |
- |
hypothetical protein JCGZ_24094 [Jatropha curcas] |
| 15 |
Hb_151281_010 |
0.1779556213 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable L-type lectin-domain containing receptor kinase S.5 [Jatropha curcas] |
| 16 |
Hb_000101_060 |
0.1789725366 |
- |
- |
- |
| 17 |
Hb_001514_100 |
0.1802624342 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 18 |
Hb_007773_030 |
0.1803208235 |
- |
- |
- |
| 19 |
Hb_002400_250 |
0.1806044827 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_002481_070 |
0.1810486816 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |