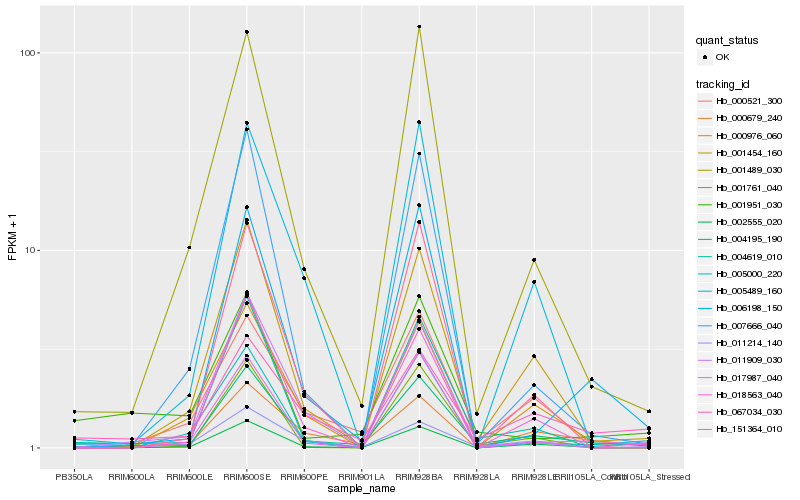
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004195_190 |
0.0 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_002555_020 |
0.1191726496 |
- |
- |
Receptor-like protein kinase [Medicago truncatula] |
| 3 |
Hb_001454_160 |
0.1419167607 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000521_300 |
0.1484438906 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 5 |
Hb_001489_030 |
0.1533632001 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 6 |
Hb_004619_010 |
0.1557367724 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 7 |
Hb_000679_240 |
0.1648223585 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 8 |
Hb_067034_030 |
0.172382301 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 9 |
Hb_017987_040 |
0.1740108991 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s11440g [Populus trichocarpa] |
| 10 |
Hb_006198_150 |
0.1767384592 |
- |
- |
- |
| 11 |
Hb_018563_040 |
0.1794188609 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_001761_040 |
0.1823404258 |
- |
- |
PREDICTED: beta-amyrin synthase-like [Jatropha curcas] |
| 13 |
Hb_011214_140 |
0.1838686147 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 14 |
Hb_007666_040 |
0.1867791731 |
- |
- |
PREDICTED: ent-kaur-16-ene synthase, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000976_060 |
0.1883950364 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_005489_160 |
0.1888666129 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 17 |
Hb_011909_030 |
0.1903203144 |
- |
- |
hypothetical protein JCGZ_00732 [Jatropha curcas] |
| 18 |
Hb_151364_010 |
0.1915752292 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 19 |
Hb_001951_030 |
0.1947941569 |
- |
- |
hypothetical protein JCGZ_04584 [Jatropha curcas] |
| 20 |
Hb_005000_220 |
0.1952734681 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |