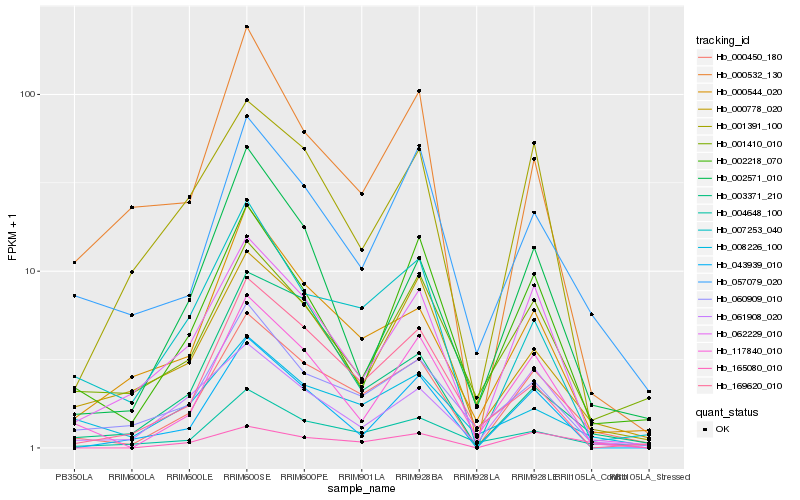
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000450_180 |
0.0 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 2 |
Hb_004648_100 |
0.153360291 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 3 |
Hb_062229_010 |
0.1541088349 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 4 |
Hb_007253_040 |
0.1596682144 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-10 isoform X1 [Jatropha curcas] |
| 5 |
Hb_057079_020 |
0.1627434852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_117840_010 |
0.1648827841 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 7 |
Hb_000544_020 |
0.1678086728 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 8 |
Hb_060909_010 |
0.1691344884 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 9 |
Hb_165080_010 |
0.1740051509 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 10 |
Hb_002218_070 |
0.1776181711 |
- |
- |
PREDICTED: myosin-binding protein 1 [Jatropha curcas] |
| 11 |
Hb_061908_020 |
0.1791211026 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 12 |
Hb_169620_010 |
0.1793779216 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 13 |
Hb_000778_020 |
0.1820865672 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 14 |
Hb_001391_100 |
0.1827938632 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 15 |
Hb_001410_010 |
0.1859388337 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP7-like [Jatropha curcas] |
| 16 |
Hb_003371_210 |
0.1862987302 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_043939_010 |
0.1871721556 |
- |
- |
hypothetical protein B456_N011100 [Gossypium raimondii] |
| 18 |
Hb_008226_100 |
0.1874757925 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 19 |
Hb_000532_130 |
0.1889829463 |
- |
- |
hypothetical protein [Anabaena sp. PCC 7108] |
| 20 |
Hb_002571_010 |
0.1894569366 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |