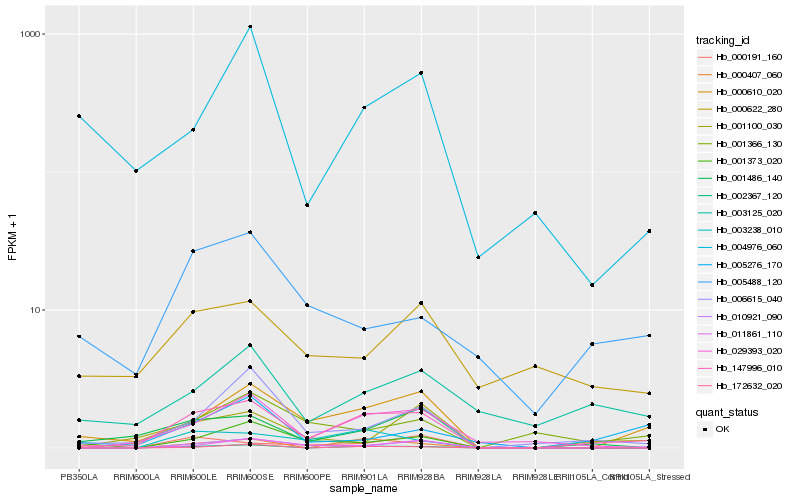
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_147996_010 |
0.0 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000610_020 |
0.2163852176 |
- |
- |
unknown [Glycine max] |
| 3 |
Hb_029393_020 |
0.2439871908 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 12-like [Jatropha curcas] |
| 4 |
Hb_010921_090 |
0.2456111105 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 5 |
Hb_001486_140 |
0.2485974525 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Vitis vinifera] |
| 6 |
Hb_002367_120 |
0.2512429983 |
- |
- |
- |
| 7 |
Hb_006615_040 |
0.2561236451 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 8 |
Hb_003238_010 |
0.2742504333 |
- |
- |
PREDICTED: uncharacterized protein LOC104880636 [Vitis vinifera] |
| 9 |
Hb_000407_060 |
0.2792756451 |
- |
- |
- |
| 10 |
Hb_001100_030 |
0.2869570935 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 11 |
Hb_001366_130 |
0.2918192164 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_003125_020 |
0.2959828092 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 13 |
Hb_011861_110 |
0.2986148625 |
- |
- |
PREDICTED: uncharacterized protein LOC105636580 [Jatropha curcas] |
| 14 |
Hb_001373_020 |
0.3051432458 |
- |
- |
- |
| 15 |
Hb_004976_060 |
0.3071064804 |
- |
- |
- |
| 16 |
Hb_005488_120 |
0.3099422281 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 17 |
Hb_172632_020 |
0.3115839231 |
- |
- |
- |
| 18 |
Hb_000191_160 |
0.3127736039 |
- |
- |
hypothetical protein EUGRSUZ_C03547 [Eucalyptus grandis] |
| 19 |
Hb_000622_280 |
0.3150979625 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005276_170 |
0.3195177467 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |