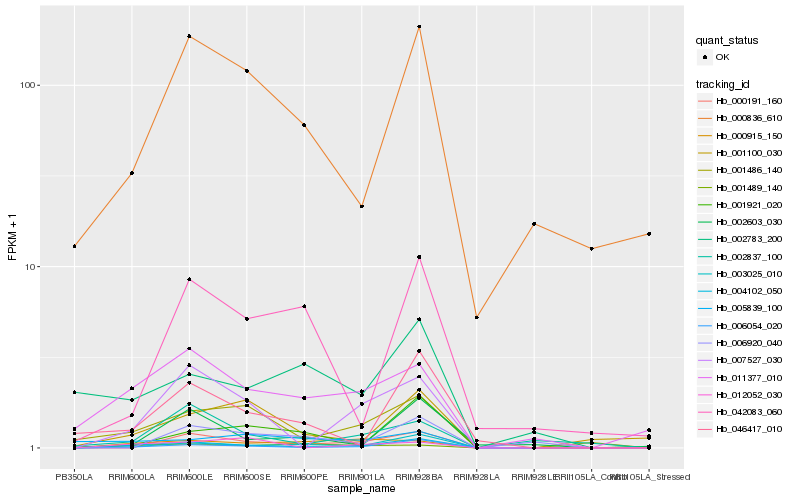
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004102_050 |
0.0 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 2 |
Hb_006054_020 |
0.1310662342 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003025_010 |
0.1956503734 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Populus euphratica] |
| 4 |
Hb_007527_030 |
0.2237894979 |
- |
- |
- |
| 5 |
Hb_001486_140 |
0.2337640494 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Vitis vinifera] |
| 6 |
Hb_000191_160 |
0.2515511639 |
- |
- |
hypothetical protein EUGRSUZ_C03547 [Eucalyptus grandis] |
| 7 |
Hb_011377_010 |
0.2569593684 |
- |
- |
PREDICTED: nudix hydrolase 12, mitochondrial-like [Populus euphratica] |
| 8 |
Hb_046417_010 |
0.2797310732 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 9 |
Hb_000915_150 |
0.2820716862 |
- |
- |
- |
| 10 |
Hb_000836_610 |
0.2841141106 |
- |
- |
stem-specific protein TSJT1-like [Jatropha curcas] |
| 11 |
Hb_002603_030 |
0.3031561687 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 12 |
Hb_001489_140 |
0.30444981 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_21694 [Jatropha curcas] |
| 13 |
Hb_002837_100 |
0.3047986105 |
- |
- |
PREDICTED: F-box protein PP2-B15-like [Jatropha curcas] |
| 14 |
Hb_001921_020 |
0.3053959216 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_001100_030 |
0.3088560413 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 16 |
Hb_012052_030 |
0.3130239536 |
- |
- |
- |
| 17 |
Hb_002783_200 |
0.3141644967 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 18 |
Hb_005839_100 |
0.3221680157 |
- |
- |
PREDICTED: APO protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 19 |
Hb_042083_060 |
0.3272441619 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 20 |
Hb_006920_040 |
0.3300799077 |
- |
- |
PREDICTED: uncharacterized protein LOC103341644 [Prunus mume] |