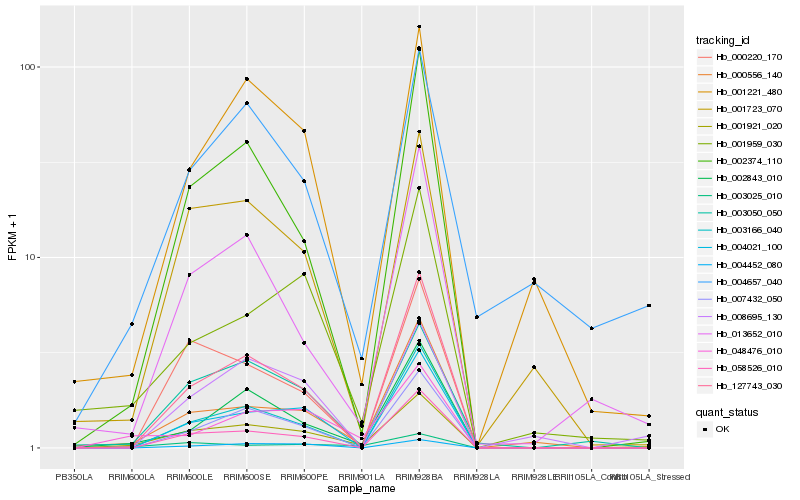
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001921_020 |
0.0 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_002374_110 |
0.1648062691 |
- |
- |
PREDICTED: uncharacterized protein LOC105638965 [Jatropha curcas] |
| 3 |
Hb_002843_010 |
0.1667917841 |
- |
- |
hypothetical protein EUGRSUZ_H04984, partial [Eucalyptus grandis] |
| 4 |
Hb_127743_030 |
0.1675199128 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_001723_070 |
0.1688964195 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 6 |
Hb_048476_010 |
0.171250251 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Citrus sinensis] |
| 7 |
Hb_007432_050 |
0.1729871276 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108 [Jatropha curcas] |
| 8 |
Hb_003050_050 |
0.1731291104 |
- |
- |
hypothetical protein JCGZ_04619 [Jatropha curcas] |
| 9 |
Hb_000556_140 |
0.1749342404 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 9 [Jatropha curcas] |
| 10 |
Hb_004021_100 |
0.175628835 |
- |
- |
- |
| 11 |
Hb_058526_010 |
0.1760472015 |
- |
- |
- |
| 12 |
Hb_001221_480 |
0.1774876921 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 13 |
Hb_003166_040 |
0.1815760081 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 14 |
Hb_003025_010 |
0.1859161475 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Populus euphratica] |
| 15 |
Hb_013652_010 |
0.187180422 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_001959_030 |
0.1888824103 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004657_040 |
0.189419309 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 18 |
Hb_004452_080 |
0.1915249181 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_008695_130 |
0.19186097 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000220_170 |
0.1920957988 |
- |
- |
- |