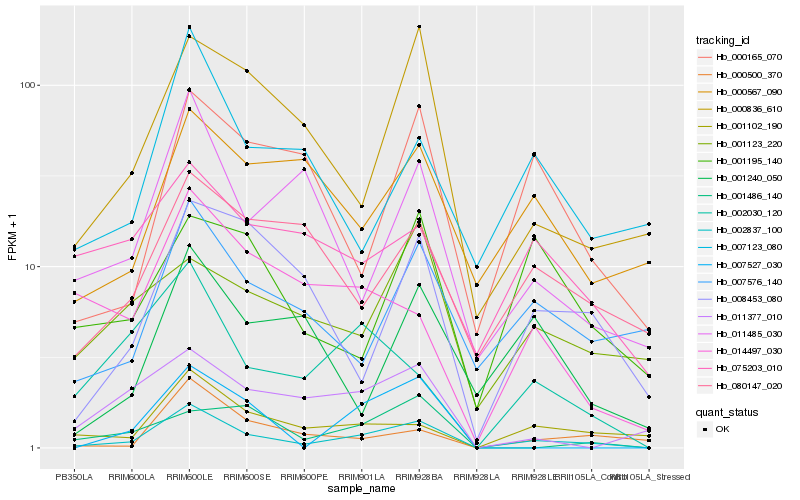
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002837_100 |
0.0 |
- |
- |
PREDICTED: F-box protein PP2-B15-like [Jatropha curcas] |
| 2 |
Hb_001102_190 |
0.2490176755 |
- |
- |
PREDICTED: uncharacterized protein LOC105112724 [Populus euphratica] |
| 3 |
Hb_000836_610 |
0.2596481727 |
- |
- |
stem-specific protein TSJT1-like [Jatropha curcas] |
| 4 |
Hb_007576_140 |
0.2625256896 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 5 |
Hb_002030_120 |
0.2649457642 |
- |
- |
- |
| 6 |
Hb_000500_370 |
0.2695891288 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 7 |
Hb_011485_030 |
0.2723600873 |
- |
- |
PREDICTED: uncharacterized protein LOC105647814 [Jatropha curcas] |
| 8 |
Hb_007527_030 |
0.2736472495 |
- |
- |
- |
| 9 |
Hb_008453_080 |
0.2760738708 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 6 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000165_070 |
0.2791625574 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |
| 11 |
Hb_007123_080 |
0.2811697584 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 12 |
Hb_011377_010 |
0.2848599212 |
- |
- |
PREDICTED: nudix hydrolase 12, mitochondrial-like [Populus euphratica] |
| 13 |
Hb_080147_020 |
0.2874427189 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 14 |
Hb_001240_050 |
0.2889735657 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Jatropha curcas] |
| 15 |
Hb_000567_090 |
0.2903972577 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_001486_140 |
0.2915107065 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Vitis vinifera] |
| 17 |
Hb_001195_140 |
0.2920972297 |
- |
- |
PREDICTED: U-box domain-containing protein 5-like [Jatropha curcas] |
| 18 |
Hb_014497_030 |
0.2923433802 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 19 |
Hb_075203_010 |
0.2928686766 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001123_220 |
0.2934089313 |
- |
- |
- |