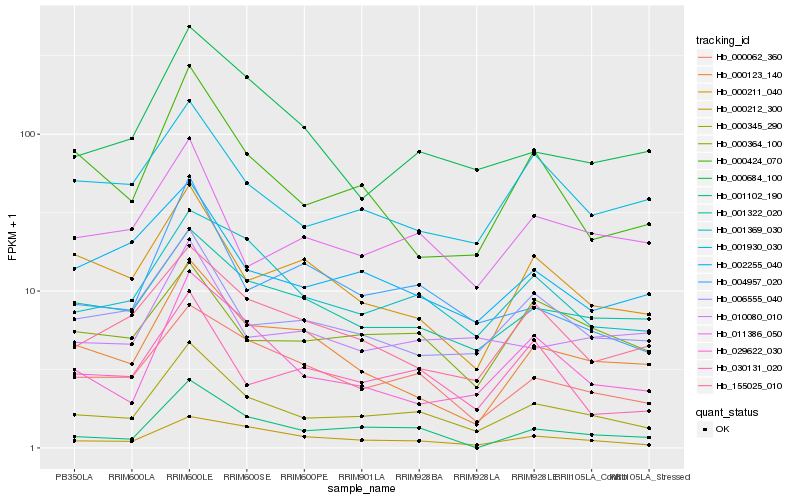
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_290 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_001369_030 |
0.1189854617 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 3 |
Hb_000062_360 |
0.1192289614 |
- |
- |
PREDICTED: DNA polymerase alpha subunit B [Jatropha curcas] |
| 4 |
Hb_000212_300 |
0.1226565132 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 5 |
Hb_001322_020 |
0.1269878985 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 6 |
Hb_006555_040 |
0.1286410154 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 7 |
Hb_029622_030 |
0.1333924894 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000684_100 |
0.1371938471 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 9 |
Hb_004957_020 |
0.1374098098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001930_030 |
0.1388073696 |
- |
- |
PREDICTED: 30S ribosomal protein S10, chloroplastic [Jatropha curcas] |
| 11 |
Hb_011386_050 |
0.1404990689 |
- |
- |
PREDICTED: malonyl-CoA-acyl carrier protein transacylase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_155025_010 |
0.1409734324 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 13 |
Hb_000424_070 |
0.141888157 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 14 |
Hb_002255_040 |
0.142879264 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 15 |
Hb_010080_010 |
0.1461122829 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 16 |
Hb_030131_020 |
0.1474733602 |
- |
- |
Carboxypeptidase B2 precursor, putative [Ricinus communis] |
| 17 |
Hb_000123_140 |
0.1477135722 |
- |
- |
PREDICTED: 21 kDa protein-like [Populus euphratica] |
| 18 |
Hb_001102_190 |
0.1480814087 |
- |
- |
PREDICTED: uncharacterized protein LOC105112724 [Populus euphratica] |
| 19 |
Hb_000364_100 |
0.148120258 |
- |
- |
PREDICTED: uncharacterized protein LOC105639739 [Jatropha curcas] |
| 20 |
Hb_000211_040 |
0.1484700255 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |