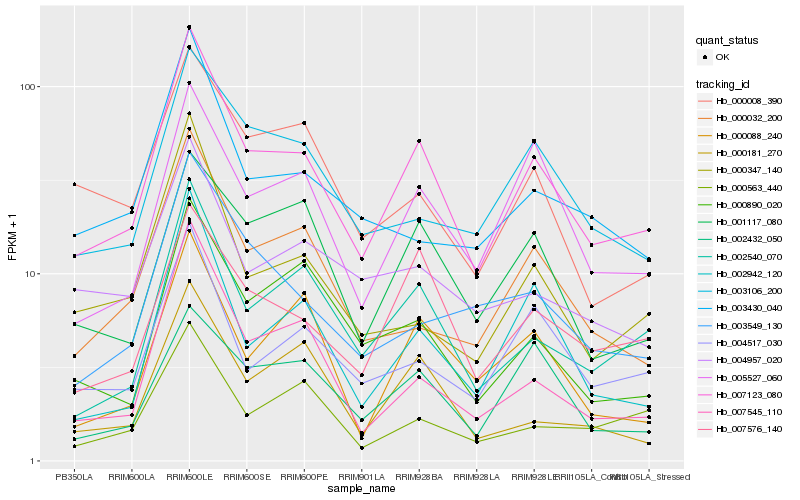
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007123_080 |
0.0 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 2 |
Hb_002540_070 |
0.1226058393 |
- |
- |
PREDICTED: protein cornichon homolog 1 [Jatropha curcas] |
| 3 |
Hb_007576_140 |
0.1359542307 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 4 |
Hb_005527_060 |
0.1360109896 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 5 |
Hb_004517_030 |
0.1377503295 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002942_120 |
0.1404898137 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 7 |
Hb_000032_200 |
0.1477167676 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 8 |
Hb_007545_110 |
0.1487255799 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_003106_200 |
0.1489392899 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001117_080 |
0.1535360608 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000890_020 |
0.1536560561 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 12 |
Hb_000347_140 |
0.1540356093 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_004957_020 |
0.1549551113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000088_240 |
0.1565918378 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000181_270 |
0.157844569 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_003549_130 |
0.1579087279 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 17 |
Hb_000008_390 |
0.1590509274 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 18 |
Hb_003430_040 |
0.1594675102 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 19 |
Hb_000563_440 |
0.1604752493 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002432_050 |
0.1615605332 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |