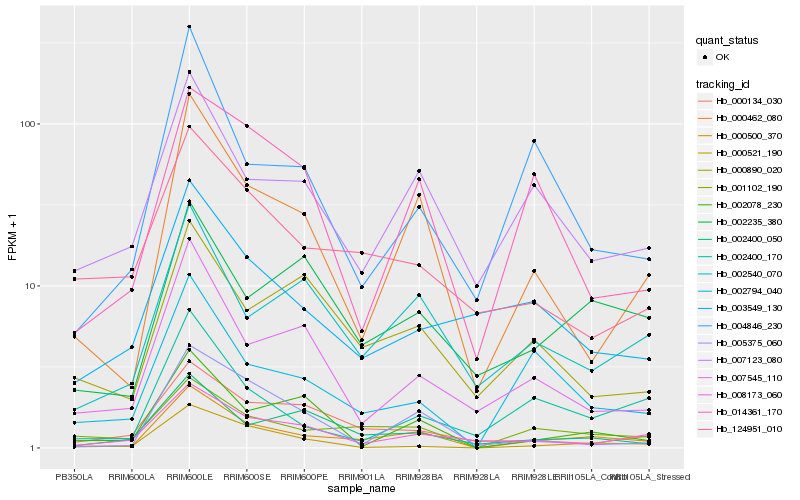
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000500_370 |
0.0 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 2 |
Hb_001102_190 |
0.167430553 |
- |
- |
PREDICTED: uncharacterized protein LOC105112724 [Populus euphratica] |
| 3 |
Hb_000462_080 |
0.1707174216 |
- |
- |
PREDICTED: quercetin 3-O-methyltransferase 1 [Jatropha curcas] |
| 4 |
Hb_002794_040 |
0.1989085577 |
- |
- |
PREDICTED: C-type lectin receptor-like tyrosine-protein kinase At1g52310 [Jatropha curcas] |
| 5 |
Hb_002540_070 |
0.2000444786 |
- |
- |
PREDICTED: protein cornichon homolog 1 [Jatropha curcas] |
| 6 |
Hb_007123_080 |
0.2014322536 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 7 |
Hb_002400_050 |
0.2114740966 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002078_230 |
0.2117271775 |
- |
- |
PREDICTED: HORMA domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_002235_380 |
0.2127363931 |
- |
- |
hypothetical protein JCGZ_19860 [Jatropha curcas] |
| 10 |
Hb_002400_170 |
0.2151974033 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 11 |
Hb_008173_060 |
0.2161879272 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 12 |
Hb_007545_110 |
0.2185051215 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_000521_190 |
0.2207782281 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_25500 [Jatropha curcas] |
| 14 |
Hb_000134_030 |
0.2252393874 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 15 |
Hb_124951_010 |
0.2265689746 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 16 |
Hb_005375_060 |
0.2272825846 |
- |
- |
PREDICTED: probably inactive receptor-like protein kinase At2g46850 [Jatropha curcas] |
| 17 |
Hb_004846_230 |
0.2275454616 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_003549_130 |
0.2308854984 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 19 |
Hb_000890_020 |
0.2310053959 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 20 |
Hb_014361_170 |
0.2331901699 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 37 [Jatropha curcas] |