| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
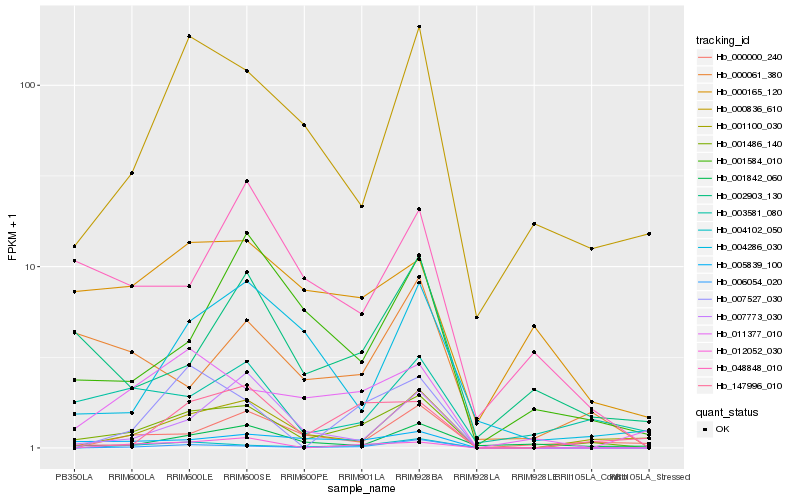
Hb_001486_140 |
0.0 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Vitis vinifera] |
| 2 |
Hb_005839_100 |
0.2284209804 |
- |
- |
PREDICTED: APO protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 3 |
Hb_001100_030 |
0.2301611962 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 4 |
Hb_004102_050 |
0.2337640494 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 5 |
Hb_147996_010 |
0.2485974525 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000836_610 |
0.2507712921 |
- |
- |
stem-specific protein TSJT1-like [Jatropha curcas] |
| 7 |
Hb_006054_020 |
0.2593509645 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003581_080 |
0.2648235477 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 9 |
Hb_012052_030 |
0.2680305519 |
- |
- |
- |
| 10 |
Hb_011377_010 |
0.2681894808 |
- |
- |
PREDICTED: nudix hydrolase 12, mitochondrial-like [Populus euphratica] |
| 11 |
Hb_007527_030 |
0.2718534985 |
- |
- |
- |
| 12 |
Hb_007773_030 |
0.2718879816 |
- |
- |
- |
| 13 |
Hb_000061_380 |
0.2739915604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001584_010 |
0.2753080109 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 15 |
Hb_002903_130 |
0.275370871 |
- |
- |
epoxide hydrolase, putative [Ricinus communis] |
| 16 |
Hb_001842_060 |
0.2799269879 |
desease resistance |
Gene Name: ABC_trans_N |
hypothetical protein JCGZ_10908 [Jatropha curcas] |
| 17 |
Hb_004286_030 |
0.2803692483 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 18 |
Hb_048848_010 |
0.2814629842 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 19 |
Hb_000165_120 |
0.2824803094 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 20 |
Hb_000000_240 |
0.282629585 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |