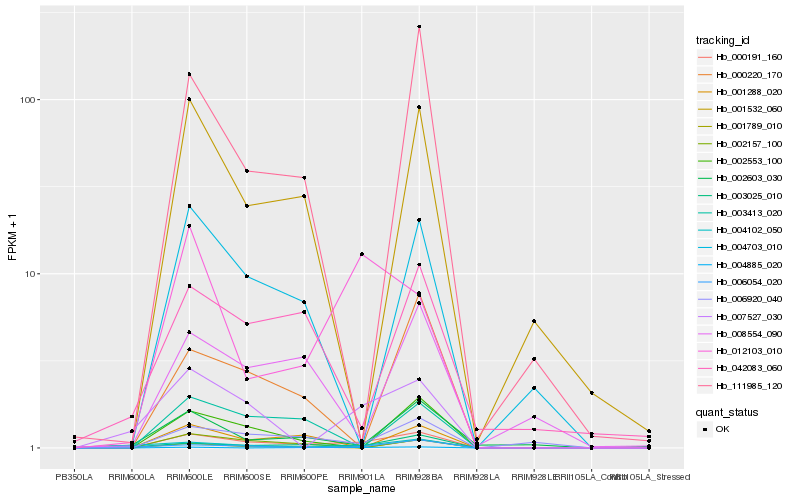
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000191_160 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_C03547 [Eucalyptus grandis] |
| 2 |
Hb_006920_040 |
0.2256826232 |
- |
- |
PREDICTED: uncharacterized protein LOC103341644 [Prunus mume] |
| 3 |
Hb_003025_010 |
0.2264502447 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Populus euphratica] |
| 4 |
Hb_006054_020 |
0.241827193 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004102_050 |
0.2515511639 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 6 |
Hb_007527_030 |
0.2612762248 |
- |
- |
- |
| 7 |
Hb_002553_100 |
0.2664133436 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor [Morus notabilis] |
| 8 |
Hb_004885_020 |
0.2742063899 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_004703_010 |
0.2786442384 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 10 |
Hb_001288_020 |
0.2818215466 |
- |
- |
- |
| 11 |
Hb_001532_060 |
0.2827138108 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_003413_020 |
0.2846128299 |
- |
- |
- |
| 13 |
Hb_008554_090 |
0.285027503 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_111985_120 |
0.2869939518 |
- |
- |
dihydroflavonol-4-reductase [Vitis rotundifolia] |
| 15 |
Hb_000220_170 |
0.2872266297 |
- |
- |
- |
| 16 |
Hb_001789_010 |
0.287377874 |
- |
- |
hypothetical protein POPTR_0015s13020g [Populus trichocarpa] |
| 17 |
Hb_042083_060 |
0.2882591059 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 18 |
Hb_002603_030 |
0.2917110371 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 19 |
Hb_012103_010 |
0.2927868889 |
- |
- |
2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [Theobroma cacao] |
| 20 |
Hb_002157_100 |
0.2956495834 |
- |
- |
- |