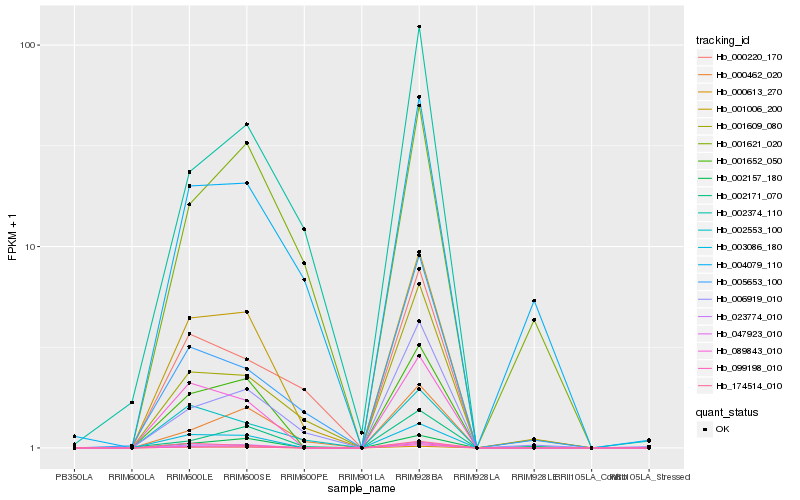
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001006_200 |
0.0 |
transcription factor |
TF Family: G2-like |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003086_180 |
0.1146262494 |
- |
- |
PREDICTED: RING-H2 finger protein ATL70-like [Pyrus x bretschneideri] |
| 3 |
Hb_001652_050 |
0.1148744614 |
- |
- |
- |
| 4 |
Hb_023774_010 |
0.1182868611 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 5 |
Hb_000613_270 |
0.1203389407 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 6 |
Hb_089843_010 |
0.1341916976 |
- |
- |
hypothetical protein JCGZ_05278 [Jatropha curcas] |
| 7 |
Hb_047923_010 |
0.1351460028 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 8 |
Hb_001609_080 |
0.1365800538 |
- |
- |
hypothetical protein POPTR_0017s05660g [Populus trichocarpa] |
| 9 |
Hb_002553_100 |
0.1462802705 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor [Morus notabilis] |
| 10 |
Hb_000462_020 |
0.1464028682 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_006919_010 |
0.1486383561 |
- |
- |
PREDICTED: putative wall-associated receptor kinase-like 16 [Vitis vinifera] |
| 12 |
Hb_004079_110 |
0.1507991011 |
- |
- |
PREDICTED: uncharacterized protein LOC100257802 [Vitis vinifera] |
| 13 |
Hb_174514_010 |
0.1527552556 |
- |
- |
flavin-containing monooxygenase family protein [Populus trichocarpa] |
| 14 |
Hb_002157_180 |
0.1560172121 |
- |
- |
separase, putative [Ricinus communis] |
| 15 |
Hb_005653_100 |
0.1574431381 |
- |
- |
hypothetical protein POPTR_0004s19580g [Populus trichocarpa] |
| 16 |
Hb_000220_170 |
0.1653340964 |
- |
- |
- |
| 17 |
Hb_001621_020 |
0.1679298427 |
- |
- |
- |
| 18 |
Hb_002374_110 |
0.1716510964 |
- |
- |
PREDICTED: uncharacterized protein LOC105638965 [Jatropha curcas] |
| 19 |
Hb_099198_010 |
0.1718535984 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At2g25790 isoform X2 [Nicotiana tomentosiformis] |
| 20 |
Hb_002171_070 |
0.171960032 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance gene-like protein ARGH30 [Populus trichocarpa] |