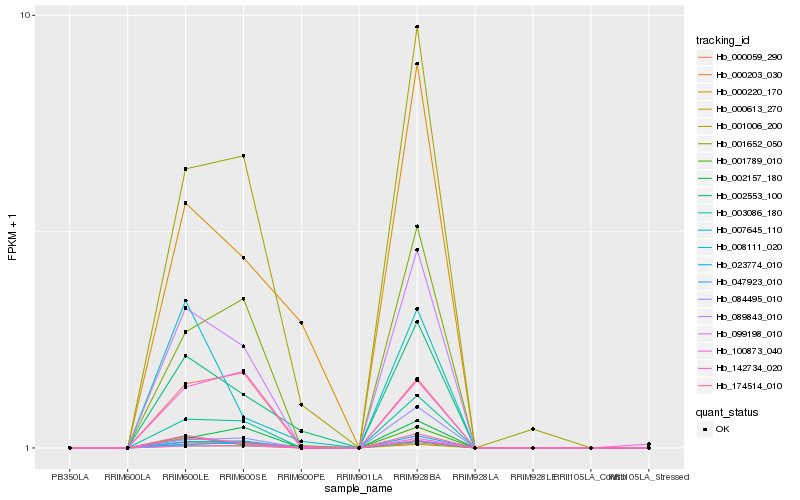
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_174514_010 |
0.0 |
- |
- |
flavin-containing monooxygenase family protein [Populus trichocarpa] |
| 2 |
Hb_089843_010 |
0.0291864368 |
- |
- |
hypothetical protein JCGZ_05278 [Jatropha curcas] |
| 3 |
Hb_003086_180 |
0.0784664207 |
- |
- |
PREDICTED: RING-H2 finger protein ATL70-like [Pyrus x bretschneideri] |
| 4 |
Hb_023774_010 |
0.1043409314 |
- |
- |
PREDICTED: uncharacterized protein LOC105635099 [Jatropha curcas] |
| 5 |
Hb_000613_270 |
0.1046121807 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 6 |
Hb_047923_010 |
0.1296197784 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 7 |
Hb_001652_050 |
0.1413367113 |
- |
- |
- |
| 8 |
Hb_001006_200 |
0.1527552556 |
transcription factor |
TF Family: G2-like |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002553_100 |
0.1548989339 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor [Morus notabilis] |
| 10 |
Hb_142734_020 |
0.1659763662 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X1 [Gossypium raimondii] |
| 11 |
Hb_000059_290 |
0.1814153842 |
- |
- |
pterin-4-alpha-carbinolamine dehydratase, putative [Ricinus communis] |
| 12 |
Hb_000203_030 |
0.1829105854 |
- |
- |
- |
| 13 |
Hb_007645_110 |
0.1907020746 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0089590 [Ricinus communis] |
| 14 |
Hb_099198_010 |
0.202976218 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At2g25790 isoform X2 [Nicotiana tomentosiformis] |
| 15 |
Hb_002157_180 |
0.205388917 |
- |
- |
separase, putative [Ricinus communis] |
| 16 |
Hb_008111_020 |
0.209302009 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 17 |
Hb_001789_010 |
0.2188300339 |
- |
- |
hypothetical protein POPTR_0015s13020g [Populus trichocarpa] |
| 18 |
Hb_100873_040 |
0.219065883 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 19 |
Hb_084495_010 |
0.2211738244 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Eucalyptus grandis] |
| 20 |
Hb_000220_170 |
0.2247514476 |
- |
- |
- |