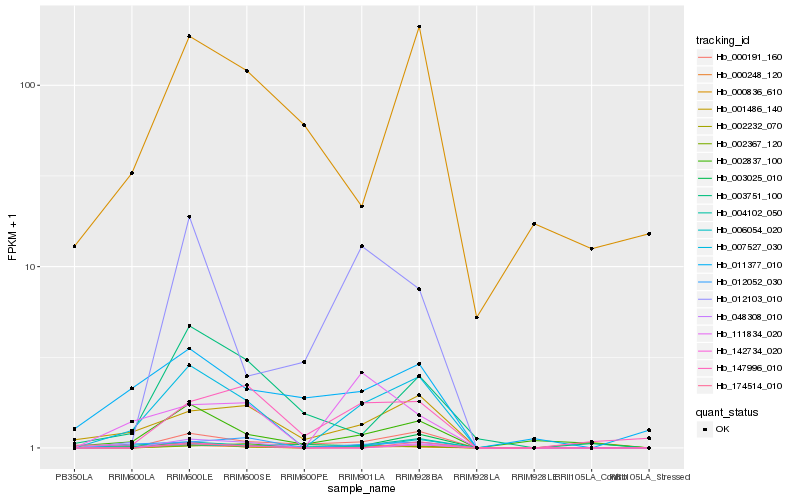
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007527_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_004102_050 |
0.2237894979 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 3 |
Hb_012052_030 |
0.2571319302 |
- |
- |
- |
| 4 |
Hb_000191_160 |
0.2612762248 |
- |
- |
hypothetical protein EUGRSUZ_C03547 [Eucalyptus grandis] |
| 5 |
Hb_006054_020 |
0.2619779058 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001486_140 |
0.2718534985 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Vitis vinifera] |
| 7 |
Hb_002837_100 |
0.2736472495 |
- |
- |
PREDICTED: F-box protein PP2-B15-like [Jatropha curcas] |
| 8 |
Hb_002232_070 |
0.3038858569 |
- |
- |
PREDICTED: putative F-box/FBD/LRR-repeat protein At4g03220 [Malus domestica] |
| 9 |
Hb_111834_020 |
0.3099089599 |
- |
- |
PREDICTED: protein phosphatase methylesterase 1 [Jatropha curcas] |
| 10 |
Hb_000248_120 |
0.3138132073 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 11 |
Hb_002367_120 |
0.3176819995 |
- |
- |
- |
| 12 |
Hb_147996_010 |
0.3224149473 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003751_100 |
0.3241025337 |
- |
- |
Amino acid permease 2 [Glycine soja] |
| 14 |
Hb_142734_020 |
0.3264753256 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X1 [Gossypium raimondii] |
| 15 |
Hb_011377_010 |
0.3278914107 |
- |
- |
PREDICTED: nudix hydrolase 12, mitochondrial-like [Populus euphratica] |
| 16 |
Hb_012103_010 |
0.3300972424 |
- |
- |
2-nonaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase [Theobroma cacao] |
| 17 |
Hb_174514_010 |
0.3412229723 |
- |
- |
flavin-containing monooxygenase family protein [Populus trichocarpa] |
| 18 |
Hb_003025_010 |
0.3467325753 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Populus euphratica] |
| 19 |
Hb_000836_610 |
0.347471081 |
- |
- |
stem-specific protein TSJT1-like [Jatropha curcas] |
| 20 |
Hb_048308_010 |
0.347837434 |
- |
- |
Peroxidase superfamily protein [Theobroma cacao] |