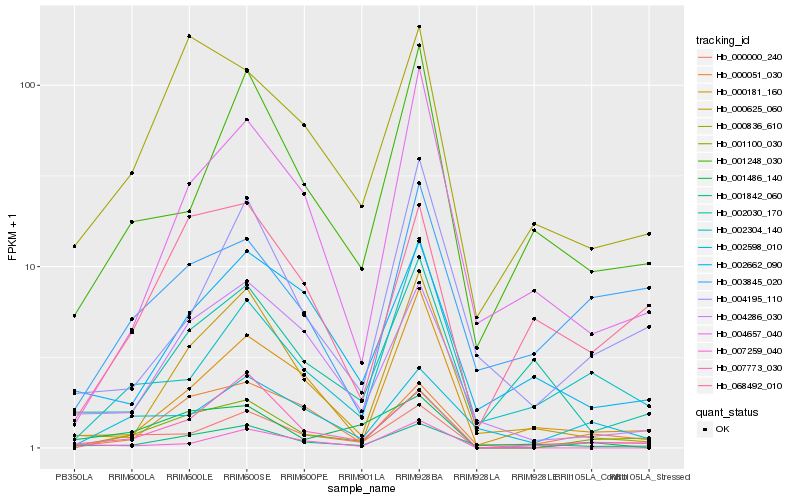
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001100_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 2 |
Hb_000000_240 |
0.1643742754 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 3 |
Hb_000051_030 |
0.1893991854 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 4 |
Hb_000836_610 |
0.1993065355 |
- |
- |
stem-specific protein TSJT1-like [Jatropha curcas] |
| 5 |
Hb_068492_010 |
0.202761074 |
- |
- |
- |
| 6 |
Hb_001842_060 |
0.2163820096 |
desease resistance |
Gene Name: ABC_trans_N |
hypothetical protein JCGZ_10908 [Jatropha curcas] |
| 7 |
Hb_004286_030 |
0.2183137143 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 8 |
Hb_003845_020 |
0.2274594446 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_001486_140 |
0.2301611962 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Vitis vinifera] |
| 10 |
Hb_002598_010 |
0.2327488897 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_001248_030 |
0.235302963 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_007259_040 |
0.2362600878 |
- |
- |
hypothetical protein VITISV_005492 [Vitis vinifera] |
| 13 |
Hb_000625_060 |
0.2434642643 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 14 |
Hb_004657_040 |
0.2438468251 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 15 |
Hb_002304_140 |
0.2459611661 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 16 |
Hb_000181_160 |
0.2464818111 |
- |
- |
PREDICTED: uncharacterized protein LOC105637575 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004195_110 |
0.2471582245 |
- |
- |
hypothetical protein JCGZ_24010 [Jatropha curcas] |
| 18 |
Hb_002662_090 |
0.2479559225 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 19 |
Hb_007773_030 |
0.2493227242 |
- |
- |
- |
| 20 |
Hb_002030_170 |
0.2521236124 |
- |
- |
PREDICTED: calcium-transporting ATPase 1-like [Jatropha curcas] |