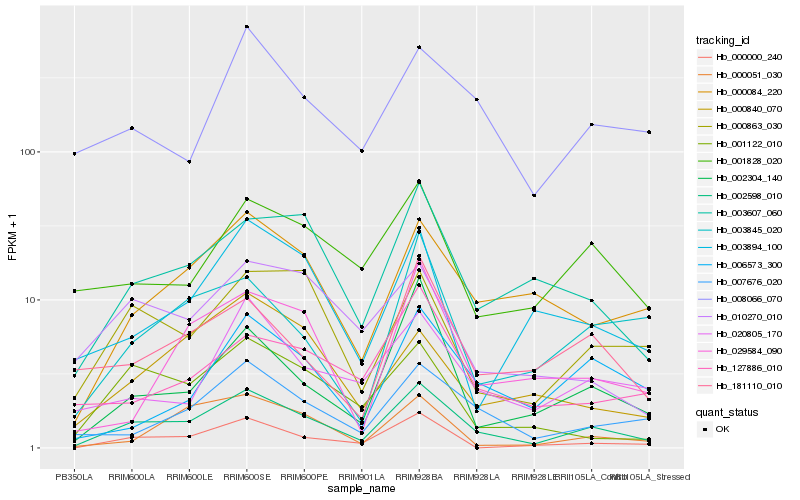
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002598_010 |
0.0 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 2 |
Hb_007676_020 |
0.1804358795 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003607_060 |
0.1824406189 |
- |
- |
PREDICTED: alcohol dehydrogenase-like [Jatropha curcas] |
| 4 |
Hb_000084_220 |
0.1831019396 |
- |
- |
PREDICTED: uncharacterized protein LOC105634692 [Jatropha curcas] |
| 5 |
Hb_000863_030 |
0.1871894947 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 6 |
Hb_000000_240 |
0.1895350006 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 7 |
Hb_181110_010 |
0.1991291955 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 8 |
Hb_020805_170 |
0.2017500943 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 9 |
Hb_002304_140 |
0.2021335786 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 10 |
Hb_001122_010 |
0.2021619103 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 11 |
Hb_006573_300 |
0.2036701297 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_001828_020 |
0.2039283712 |
- |
- |
hypothetical protein CISIN_1g010132mg [Citrus sinensis] |
| 13 |
Hb_008066_070 |
0.2053061917 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 14 |
Hb_003845_020 |
0.2085131706 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_029584_090 |
0.2110167591 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 16 |
Hb_000051_030 |
0.2111638107 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 17 |
Hb_000840_070 |
0.2138981294 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_003894_100 |
0.2153668882 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 19 |
Hb_010270_010 |
0.215494033 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_127886_010 |
0.2167259432 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |