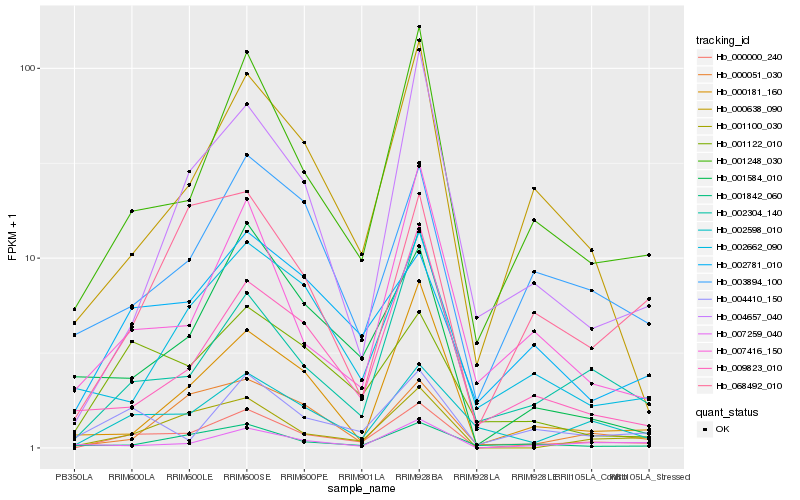
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_240 |
0.0 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 2 |
Hb_001248_030 |
0.1632824458 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_001100_030 |
0.1643742754 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 4 |
Hb_002598_010 |
0.1895350006 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 5 |
Hb_002304_140 |
0.1919806101 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 6 |
Hb_000638_090 |
0.2073887696 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_002781_010 |
0.2087124574 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_004410_150 |
0.2108624318 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 9 |
Hb_001122_010 |
0.2147917543 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 10 |
Hb_007259_040 |
0.2148937141 |
- |
- |
hypothetical protein VITISV_005492 [Vitis vinifera] |
| 11 |
Hb_003894_100 |
0.2164017038 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 12 |
Hb_007416_150 |
0.2179751483 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 13 |
Hb_001842_060 |
0.2190486621 |
desease resistance |
Gene Name: ABC_trans_N |
hypothetical protein JCGZ_10908 [Jatropha curcas] |
| 14 |
Hb_000051_030 |
0.2190702329 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 15 |
Hb_068492_010 |
0.2216565454 |
- |
- |
- |
| 16 |
Hb_001584_010 |
0.2217948602 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 17 |
Hb_004657_040 |
0.2232805672 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 18 |
Hb_002662_090 |
0.2243337936 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 19 |
Hb_009823_010 |
0.2263163756 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 20 |
Hb_000181_160 |
0.2284311725 |
- |
- |
PREDICTED: uncharacterized protein LOC105637575 isoform X2 [Jatropha curcas] |