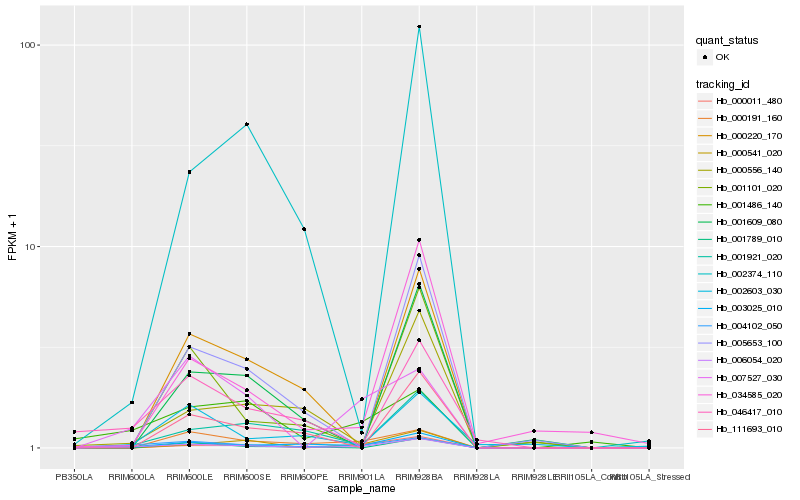
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006054_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004102_050 |
0.1310662342 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 3 |
Hb_003025_010 |
0.1500902365 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Populus euphratica] |
| 4 |
Hb_001921_020 |
0.2374757154 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000191_160 |
0.241827193 |
- |
- |
hypothetical protein EUGRSUZ_C03547 [Eucalyptus grandis] |
| 6 |
Hb_046417_010 |
0.2539199252 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 7 |
Hb_001486_140 |
0.2593509645 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Vitis vinifera] |
| 8 |
Hb_007527_030 |
0.2619779058 |
- |
- |
- |
| 9 |
Hb_005653_100 |
0.2638015329 |
- |
- |
hypothetical protein POPTR_0004s19580g [Populus trichocarpa] |
| 10 |
Hb_000011_480 |
0.2676140791 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000541_020 |
0.2717195009 |
- |
- |
- |
| 12 |
Hb_000220_170 |
0.273047295 |
- |
- |
- |
| 13 |
Hb_111693_010 |
0.2742417478 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 14 |
Hb_034585_020 |
0.2748539585 |
- |
- |
- |
| 15 |
Hb_001101_020 |
0.2771656121 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 16 |
Hb_002374_110 |
0.2779595567 |
- |
- |
PREDICTED: uncharacterized protein LOC105638965 [Jatropha curcas] |
| 17 |
Hb_002603_030 |
0.2783087418 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 18 |
Hb_000556_140 |
0.2801331028 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 9 [Jatropha curcas] |
| 19 |
Hb_001789_010 |
0.2801833076 |
- |
- |
hypothetical protein POPTR_0015s13020g [Populus trichocarpa] |
| 20 |
Hb_001609_080 |
0.2802438844 |
- |
- |
hypothetical protein POPTR_0017s05660g [Populus trichocarpa] |