| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
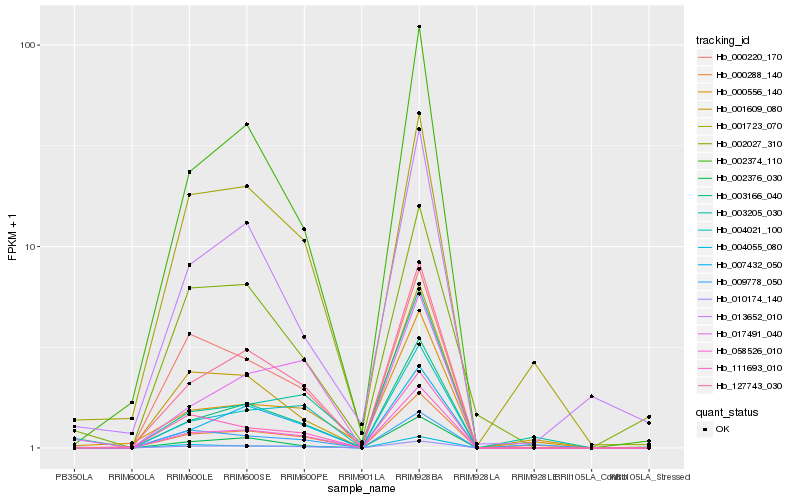
Hb_010174_140 |
0.0 |
- |
- |
Alpha-L-fucosidase 2 precursor, putative [Ricinus communis] |
| 2 |
Hb_058526_010 |
0.1770982733 |
- |
- |
- |
| 3 |
Hb_003166_040 |
0.1842531802 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 4 |
Hb_127743_030 |
0.1844781462 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_017491_040 |
0.1883255792 |
- |
- |
PREDICTED: uncharacterized protein LOC105631348 [Jatropha curcas] |
| 6 |
Hb_000220_170 |
0.1898504897 |
- |
- |
- |
| 7 |
Hb_002374_110 |
0.1899045533 |
- |
- |
PREDICTED: uncharacterized protein LOC105638965 [Jatropha curcas] |
| 8 |
Hb_002376_030 |
0.1912095054 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_111693_010 |
0.1914224574 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 10 |
Hb_004055_080 |
0.1933475585 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 11 |
Hb_002027_310 |
0.1944799236 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007432_050 |
0.1968850416 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB108 [Jatropha curcas] |
| 13 |
Hb_001723_070 |
0.1971982276 |
- |
- |
PREDICTED: polygalacturonase At1g48100 [Jatropha curcas] |
| 14 |
Hb_004021_100 |
0.2011696429 |
- |
- |
- |
| 15 |
Hb_000288_140 |
0.2014210207 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 35 [Jatropha curcas] |
| 16 |
Hb_013652_010 |
0.2053798171 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_001609_080 |
0.2059181442 |
- |
- |
hypothetical protein POPTR_0017s05660g [Populus trichocarpa] |
| 18 |
Hb_000556_140 |
0.2061178625 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 9 [Jatropha curcas] |
| 19 |
Hb_003205_030 |
0.2097586488 |
- |
- |
Pectinesterase inhibitor, putative [Ricinus communis] |
| 20 |
Hb_009778_050 |
0.2098749017 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |