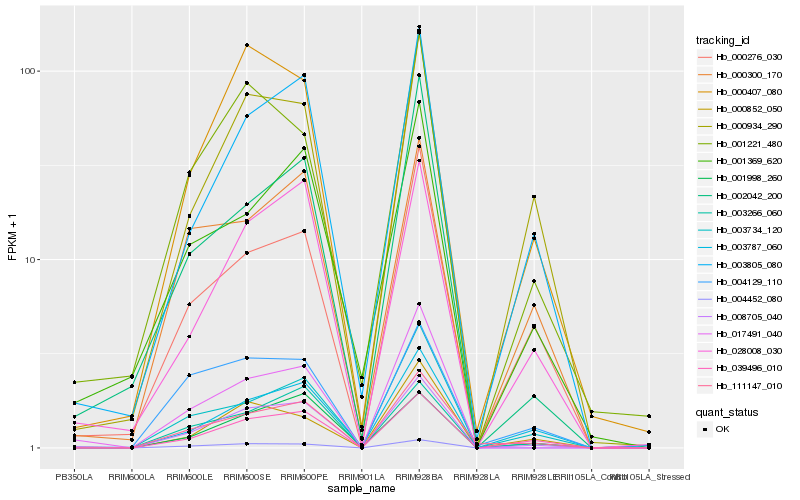
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008705_040 |
0.0 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 2 |
Hb_039496_010 |
0.1052270441 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 3 |
Hb_004452_080 |
0.1108214987 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 4 |
Hb_028008_030 |
0.1153421159 |
- |
- |
probable xyloglucan endotransglucosylase/hydrolase protein 23 precursor [Malus domestica] |
| 5 |
Hb_003805_080 |
0.1160055758 |
- |
- |
PREDICTED: isoflavone reductase-like protein [Jatropha curcas] |
| 6 |
Hb_111147_010 |
0.1182473279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_017491_040 |
0.1185497876 |
- |
- |
PREDICTED: uncharacterized protein LOC105631348 [Jatropha curcas] |
| 8 |
Hb_003787_060 |
0.1330990511 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 9 |
Hb_001998_260 |
0.1352974815 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_003266_060 |
0.1383621447 |
- |
- |
PREDICTED: basic 7S globulin-like [Populus euphratica] |
| 11 |
Hb_000407_080 |
0.1395491137 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase [Hevea brasiliensis] |
| 12 |
Hb_000934_290 |
0.1409245883 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003734_120 |
0.1420713418 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002042_200 |
0.1426895695 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_001221_480 |
0.146279028 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_000276_030 |
0.1473058643 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 17 |
Hb_001369_620 |
0.1474248788 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 18 |
Hb_000852_050 |
0.1486602215 |
- |
- |
hypothetical protein POPTR_0018s02450g [Populus trichocarpa] |
| 19 |
Hb_004129_110 |
0.1504413861 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 20 |
Hb_000300_170 |
0.150781243 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |