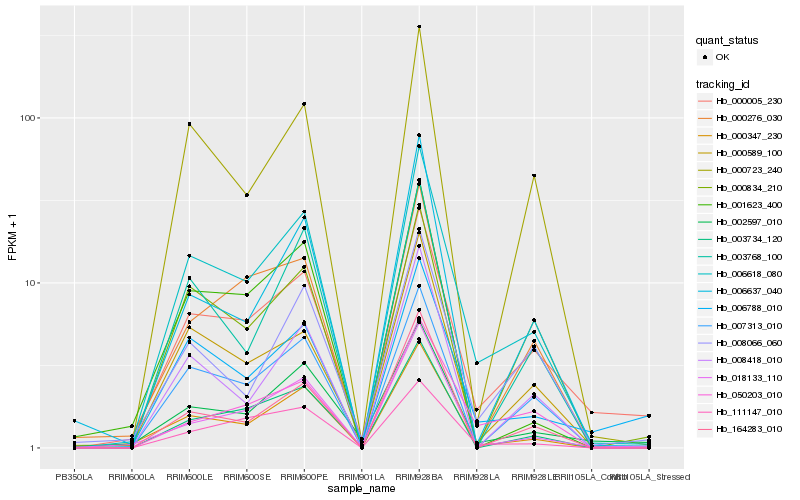
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006618_080 |
0.0 |
- |
- |
PREDICTED: non-specific lipid-transfer protein [Jatropha curcas] |
| 2 |
Hb_000347_230 |
0.0980583886 |
- |
- |
inter-alpha-trypsin inhibitor heavy chain, putative [Ricinus communis] |
| 3 |
Hb_000005_230 |
0.1169551954 |
- |
- |
hypothetical protein POPTR_0009s01160g [Populus trichocarpa] |
| 4 |
Hb_007313_010 |
0.1186447742 |
- |
- |
hypothetical protein JCGZ_23398 [Jatropha curcas] |
| 5 |
Hb_000723_240 |
0.1268049239 |
- |
- |
phenylalanine ammonia-lyase 2 [Manihot esculenta] |
| 6 |
Hb_000589_100 |
0.1295596657 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_003734_120 |
0.1313519363 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 8 |
Hb_006788_010 |
0.1315331757 |
- |
- |
PREDICTED: calcium-transporting ATPase 12, plasma membrane-type [Jatropha curcas] |
| 9 |
Hb_111147_010 |
0.1339001574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003768_100 |
0.134203276 |
- |
- |
hypothetical protein JCGZ_19930 [Jatropha curcas] |
| 11 |
Hb_018133_110 |
0.1391249516 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_008418_010 |
0.1397630435 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 13 |
Hb_050203_010 |
0.1454610576 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 14 |
Hb_001623_400 |
0.1458518697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_164283_010 |
0.1459569111 |
- |
- |
hypothetical protein RCOM_0725500 [Ricinus communis] |
| 16 |
Hb_002597_010 |
0.1476867493 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 17 |
Hb_000834_210 |
0.1479510212 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_008066_060 |
0.1484120689 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 19 |
Hb_000276_030 |
0.1511697964 |
- |
- |
dimethylaniline monooxygenase, putative [Ricinus communis] |
| 20 |
Hb_006637_040 |
0.1514713146 |
- |
- |
phenylalanine ammonia-lyase 2 [Manihot esculenta] |