| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
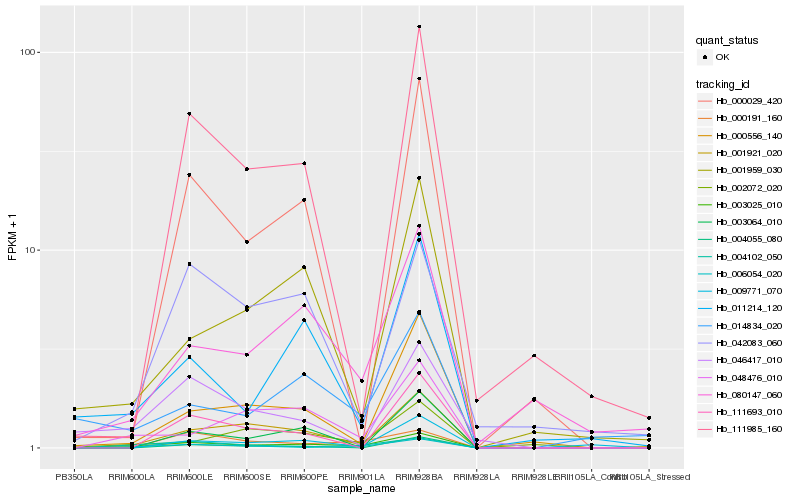
Hb_003025_010 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Populus euphratica] |
| 2 |
Hb_006054_020 |
0.1500902365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001921_020 |
0.1859161475 |
desease resistance |
Gene Name: LRR_8 |
PREDICTED: disease resistance protein RPM1-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_004102_050 |
0.1956503734 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 5 |
Hb_048476_010 |
0.2169838684 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Citrus sinensis] |
| 6 |
Hb_080147_060 |
0.2192027991 |
- |
- |
PREDICTED: cyclin-dependent kinase F-1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000191_160 |
0.2264502447 |
- |
- |
hypothetical protein EUGRSUZ_C03547 [Eucalyptus grandis] |
| 8 |
Hb_000556_140 |
0.2389096986 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 9 [Jatropha curcas] |
| 9 |
Hb_000029_420 |
0.243687512 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 10 |
Hb_111985_160 |
0.2445117217 |
- |
- |
hypothetical protein B456_010G008900 [Gossypium raimondii] |
| 11 |
Hb_004055_080 |
0.249956076 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 12 |
Hb_011214_120 |
0.2505993435 |
- |
- |
PREDICTED: uncharacterized protein LOC105636139 [Jatropha curcas] |
| 13 |
Hb_111693_010 |
0.2512590286 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 14 |
Hb_042083_060 |
0.2522624123 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g45780 [Jatropha curcas] |
| 15 |
Hb_001959_030 |
0.2526551112 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002072_020 |
0.2534026183 |
- |
- |
hypothetical protein JCGZ_16057 [Jatropha curcas] |
| 17 |
Hb_009771_070 |
0.2536273217 |
- |
- |
hypothetical protein POPTR_0016s12140g [Populus trichocarpa] |
| 18 |
Hb_014834_020 |
0.2544465713 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_046417_010 |
0.2548282466 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 20 |
Hb_003064_010 |
0.2562655866 |
- |
- |
Senescence-related gene 1 [Theobroma cacao] |