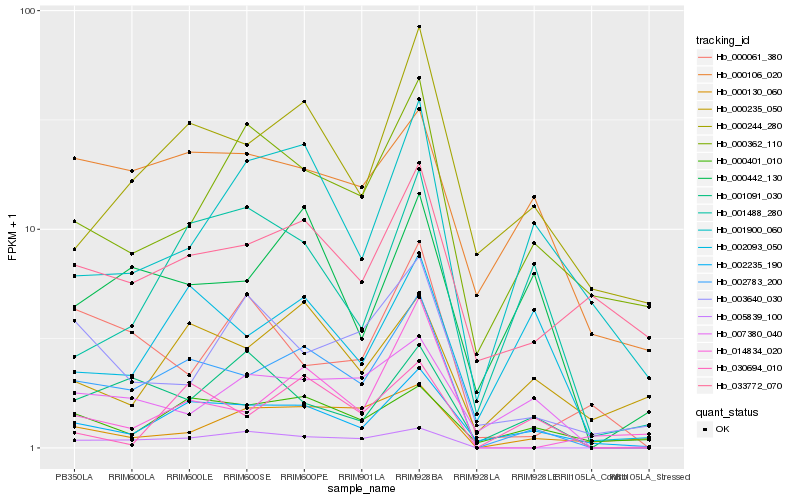
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_200 |
0.0 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 2 |
Hb_002235_190 |
0.1795900111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005839_100 |
0.1836050428 |
- |
- |
PREDICTED: APO protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 4 |
Hb_000244_280 |
0.2204839481 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000401_010 |
0.2277604976 |
- |
- |
SAB, putative [Ricinus communis] |
| 6 |
Hb_007380_040 |
0.2322796525 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 7 |
Hb_000442_130 |
0.2338050375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_030694_010 |
0.2345983708 |
- |
- |
PREDICTED: endoglucanase 25-like [Solanum tuberosum] |
| 9 |
Hb_014834_020 |
0.2374786198 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_002093_050 |
0.2418230502 |
- |
- |
hypothetical protein POPTR_0008s02940g [Populus trichocarpa] |
| 11 |
Hb_000130_060 |
0.2452691053 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 12 |
Hb_000106_020 |
0.2481045805 |
- |
- |
PREDICTED: protein TOPLESS isoform X1 [Jatropha curcas] |
| 13 |
Hb_033772_070 |
0.2507937307 |
- |
- |
PREDICTED: uncharacterized protein LOC105633681 [Jatropha curcas] |
| 14 |
Hb_003640_030 |
0.2510711432 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000362_110 |
0.2522033775 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 16 |
Hb_001488_280 |
0.2526488509 |
- |
- |
hypothetical protein JCGZ_19947 [Jatropha curcas] |
| 17 |
Hb_000235_050 |
0.2529330625 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001091_030 |
0.2532238764 |
- |
- |
hypothetical protein AALP_AA1G173500 [Arabis alpina] |
| 19 |
Hb_001900_060 |
0.2552046472 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 20 |
Hb_000061_380 |
0.2575829006 |
- |
- |
conserved hypothetical protein [Ricinus communis] |