| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
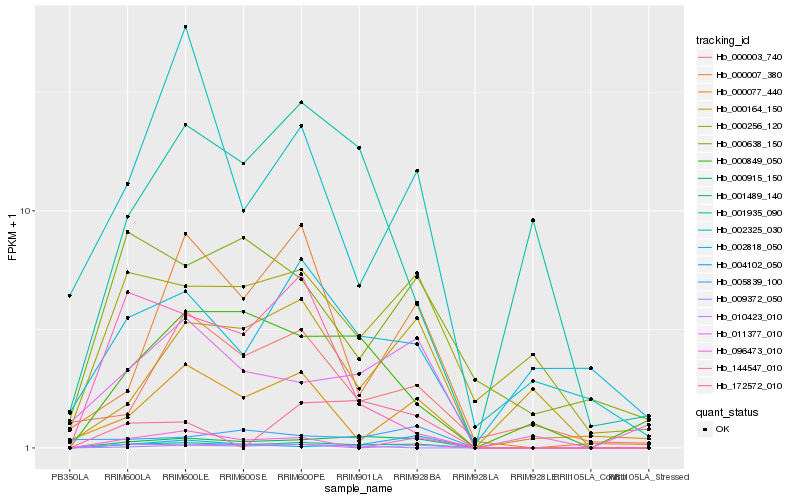
Hb_001489_140 |
0.0 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_21694 [Jatropha curcas] |
| 2 |
Hb_000915_150 |
0.1314854984 |
- |
- |
- |
| 3 |
Hb_011377_010 |
0.2456287227 |
- |
- |
PREDICTED: nudix hydrolase 12, mitochondrial-like [Populus euphratica] |
| 4 |
Hb_000256_120 |
0.2548659847 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 [Jatropha curcas] |
| 5 |
Hb_000164_150 |
0.2653309183 |
- |
- |
serine/threonine-specific protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000849_050 |
0.266724759 |
- |
- |
hypothetical protein VITISV_010852 [Vitis vinifera] |
| 7 |
Hb_096473_010 |
0.2721157367 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 8 |
Hb_000007_380 |
0.2728765308 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 9 |
Hb_010423_010 |
0.2893277927 |
- |
- |
- |
| 10 |
Hb_000003_740 |
0.2897108225 |
- |
- |
PREDICTED: uncharacterized protein LOC105631539 [Jatropha curcas] |
| 11 |
Hb_005839_100 |
0.2920732149 |
- |
- |
PREDICTED: APO protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 12 |
Hb_000638_150 |
0.2937397639 |
- |
- |
PREDICTED: U-box domain-containing protein 26-like [Jatropha curcas] |
| 13 |
Hb_001935_090 |
0.2999457029 |
- |
- |
PREDICTED: uncharacterized protein LOC105643180 isoform X1 [Jatropha curcas] |
| 14 |
Hb_172572_010 |
0.3016635122 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 15 |
Hb_002325_030 |
0.3024200013 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 2 [Jatropha curcas] |
| 16 |
Hb_000077_440 |
0.3033928977 |
- |
- |
PREDICTED: ELMO domain-containing protein A isoform X1 [Jatropha curcas] |
| 17 |
Hb_004102_050 |
0.30444981 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 18 |
Hb_144547_010 |
0.3060143811 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 19 |
Hb_002818_050 |
0.307070646 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 20 |
Hb_009372_050 |
0.3097408237 |
- |
- |
unnamed protein product [Vitis vinifera] |