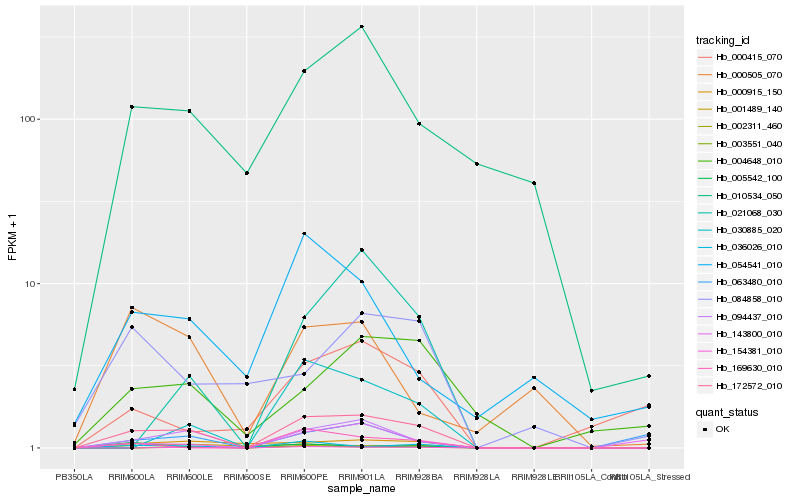
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172572_010 |
0.0 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 2 |
Hb_094437_010 |
0.1794104849 |
- |
- |
- |
| 3 |
Hb_036026_010 |
0.2615243292 |
transcription factor |
TF Family: FAR1 |
FAR-RED IMPAIRED RESPONSE [Medicago truncatula] |
| 4 |
Hb_000915_150 |
0.2758406416 |
- |
- |
- |
| 5 |
Hb_154381_010 |
0.2821981319 |
- |
- |
- |
| 6 |
Hb_003551_040 |
0.2847135198 |
- |
- |
PREDICTED: uncharacterized protein LOC103331076 [Prunus mume] |
| 7 |
Hb_010534_050 |
0.2985839434 |
- |
- |
PREDICTED: 40S ribosomal protein S7 [Jatropha curcas] |
| 8 |
Hb_001489_140 |
0.3016635122 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_21694 [Jatropha curcas] |
| 9 |
Hb_169630_010 |
0.3051209789 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 10 |
Hb_030885_020 |
0.3053889686 |
- |
- |
- |
| 11 |
Hb_004648_010 |
0.3100086662 |
- |
- |
UDP-galactose/UDP-glucose transporter 4 [Morus notabilis] |
| 12 |
Hb_143800_010 |
0.3175771976 |
- |
- |
hypothetical protein POPTR_0010s21410g, partial [Populus trichocarpa] |
| 13 |
Hb_002311_460 |
0.3231854683 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_063480_010 |
0.3295560372 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 1-like isoform X2 [Pyrus x bretschneideri] |
| 15 |
Hb_084858_010 |
0.3314547155 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 16 |
Hb_021068_030 |
0.335199435 |
transcription factor |
TF Family: M-type |
hypothetical protein B456_010G032700 [Gossypium raimondii] |
| 17 |
Hb_000505_070 |
0.3367407777 |
- |
- |
PREDICTED: heparanase-like protein 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000415_070 |
0.3378598553 |
- |
- |
- |
| 19 |
Hb_054541_010 |
0.3405184709 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 20 |
Hb_005542_100 |
0.3466141576 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0013s03910g [Populus trichocarpa] |