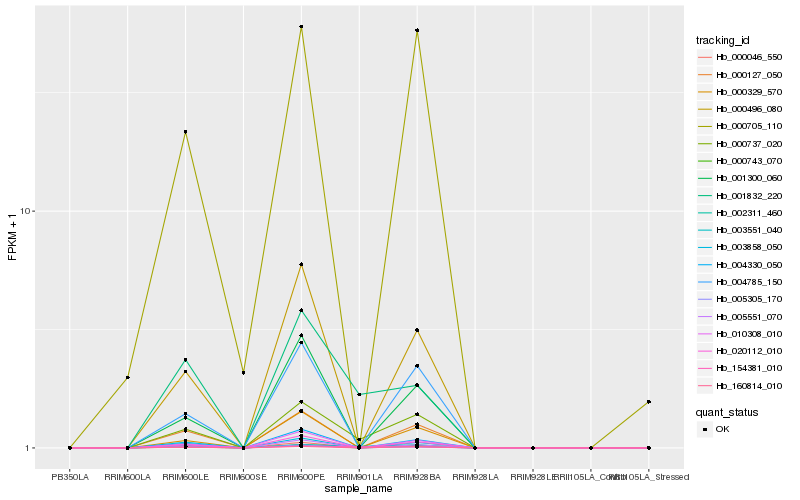
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000737_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_002311_460 |
0.1368456226 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_001832_220 |
0.1803937125 |
- |
- |
- |
| 4 |
Hb_000743_070 |
0.1896693487 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 5 |
Hb_000127_050 |
0.1934360416 |
- |
- |
- |
| 6 |
Hb_160814_010 |
0.198673172 |
- |
- |
PREDICTED: probable sulfate transporter 3.5 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004785_150 |
0.2002052861 |
- |
- |
hypothetical protein JCGZ_25207 [Jatropha curcas] |
| 8 |
Hb_020112_010 |
0.2032854462 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000496_080 |
0.2104267778 |
- |
- |
Proteinase inhibitor, putative [Ricinus communis] |
| 10 |
Hb_010308_010 |
0.2109390781 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 11 |
Hb_004330_050 |
0.2117647246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000329_570 |
0.2121039784 |
- |
- |
- |
| 13 |
Hb_000046_550 |
0.2134972074 |
- |
- |
PREDICTED: protein ALWAYS EARLY 3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003858_050 |
0.2155568877 |
- |
- |
hypothetical protein PRUPE_ppa021882mg [Prunus persica] |
| 15 |
Hb_003551_040 |
0.2170751055 |
- |
- |
PREDICTED: uncharacterized protein LOC103331076 [Prunus mume] |
| 16 |
Hb_005305_170 |
0.2180569856 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 8 [Jatropha curcas] |
| 17 |
Hb_154381_010 |
0.2183525267 |
- |
- |
- |
| 18 |
Hb_001300_060 |
0.2217435859 |
- |
- |
PREDICTED: monothiol glutaredoxin-S6-like [Jatropha curcas] |
| 19 |
Hb_000705_110 |
0.2221373729 |
- |
- |
hypothetical protein POPTR_0005s09220g [Populus trichocarpa] |
| 20 |
Hb_005551_070 |
0.2228570439 |
- |
- |
PREDICTED: uncharacterized protein LOC105635822 isoform X1 [Jatropha curcas] |